D N A b i n d i n g a n d DIMERISATION PROPERTIES OF THE TRANSCRIPTION FACTOR COUP-TF II
Alison Jane Butler
Thesis presented in partial fulfilm ent of the degree of Doctor of Philosophy at the University of London
A ugust 1995
Molecular Endocrinology Laboratory Im perial Cancer Research Fund Lincoln's In n Fields
L o n d o n
D epartm ent of M olecular Pathology U niversity College London
All rights reserved
INFORMATION TO ALL USERS
The quality of this reproduction is dependent upon the quality of the copy submitted.
In the unlikely event that the author did not send a complete manuscript
and there are missing pages, th ese will be noted. Also, if material had to be removed, a note will indicate the deletion.
uest.
ProQuest 10018647
Published by ProQuest LLC(2016). Copyright of the Dissertation is held by the Author.
All rights reserved.
This work is protected against unauthorized copying under Title 17, United States Code. Microform Edition © ProQuest LLC.
ProQuest LLC
789 East Eisenhower Parkway P.O. Box 1346
The chicken ovalbum in upstream transcription factors (COUP-TF I and II) are m em bers of the nuclear receptor family. These proteins have no identified ligand and can repress the transcriptional activity of a num ber of other nuclear receptors including the receptors for retinoic acid (RAR and RXR), thyroid horm one (TR) and oestrogen (ER). Analysis of COUP-TF protein revealed that its expression can be regulated by both retinoic acid and oestrogen indicating that COUP-TF m ediated repression in vivo m ay be subject to horm onal controls.
COUP-TF II w as show n to bind DNA in vitro either as a hom odim er or as heterodim er w ith RXRa or TRp. To establish w hether COUP-TF II could form hom odim ers and heterodim ers in vivo the dim érisation
p roperties of this orphan receptor w ere investigated in transfected cells using a tw o hybrid assay. In conditions w here COUP-TF II hom odim ers and
R X R a/R A R a heterodim ers w ere form ed heterodim ers betw een COUP-TF II and RXRa w ere not detected. M oreover, no interaction betw een COUP-TF II and RXRa could be detected on DNA in intact cells. Similarly, COUP-TF II hom odim ers and R X R a/TR p heterodim ers w ere favoured over COUP-TF II/T R p heterodim ers. These results suggest th at the form ation of
ACKNOWLEDGMENTS
Firstly I w ould like to thank Malcolm Parker for accepting m e into his lab and for the guidance and support he has provided over the last four years. I w ould also like to thank past and present m em bers of the M olecular Endocrinology Laboratory for their ideas, support and tolerance. In particular I w ould like to thank Roger White, Sue H oare and A nna Florence for advice and help. Thanks are also due to all those w ho kindly provided reagents and protocols and to the staff of the oligonucleotide and peptide synthesis
services and the photography and com puting departm ents.
Title...1
A b stract... 3
A ck n o w led g m e n ts...4
C ontents...5
A b b rev iatio n s... 13
C hapter 1 Introduction Introduction...17
The identification of intracellular horm one recep to rs...17
N uclear localisation...19
The nuclear receptor superfam ily... 20
COU P-TF...22
O ther orphan receptors...23
N uclear receptor response elem ents...24
N uclear receptor structure...29
DNA binding ... 29
D im é ris a tio n ... 36
Ligand B in d in g ... 38
T ranscriptional activation d o m a in s...39
T ranscriptional R egulation...41
Basal transcription m ediated by RNA polym erase II...41
A ctivated tran scrip tio n ...43
T ranscriptional rep ressio n ...50
Passive rep ressio n ...50
Active rep re ssio n ...52
C hapter 2 M aterials and M ethods M aterials...55
Chem icals and solvents...55
Radiochemicals...56
E n zy m es...56
M isc e llan e o u s...57
Antibodies and tissu es... 58
Buffers... 58
Bacterial m edia and agar... 62
M e th o d s... 64
Storage of bacteria... 64
Preparation of com petent b acteria...64
T ransform ation of com petent b acteria...64
P reparation of plasm id D N A ...65
Small scale preparation of plasm id DNA (mini prep)...65
Large scale preparation of plasm id D N A ...65
DNA m anipulation and subcloning...66
Restriction endonuclease d ig estio n ...66
Agarose gel electrophoresis... 66
Purification of DNA fragm ents from agarose gels...67
O ligonucleotide kinasing and annealing... 67
P reparation of v ecto rs...67
L igations...68
Polym erase chain reactio n ... 68
DNA sequencing... 68
D enaturing gel electrophoresis...69
Exonuclease III and m ung bean nuclease deletion of DNA...69
DNA screening of cDNA lib rary ...70
End labelling of oligonucleotides... 72
In vitro protein analysis...72
In vitro pro tein synthesis...72
SDS polyacrylam ide gel electrophoresis...72
W estern blotting... 73
Gel shift assay...74
D eterm ination of protein concentration...75
G eneration of polyclonal antibodies...75
Im m u n o p ré c ip ita tio n ...76
Cell culture m e th o d s...77
M aintenance of cell stocks...77
Storage of cells ...77
C harcoal treatm ent of se ru m ...78
T ransient tran sfectio n ...78
Calcium phosphate precipitation- HBS m eth o d ... 78
Luciferase activity assay ...80
CAT activity a ssa y ... 81
P-galactosidase assay ... 81
Preparation of whole cell extracts...82
C hapter 3 A nalysis of COUP-TF expression Introduction...84
Sequencing of a full length COUP-TF II cDNA...84
G eneration of polyclonal antibodies to COUP-TF...87
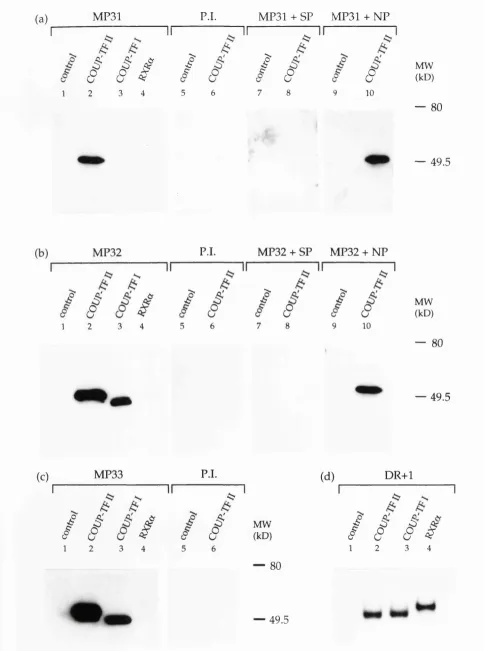
Im m unoprécipitation using MP31, MP32 and MP33 antisera... 87
W estern blot analysis of the antisera MP31, MP32 and M P33... 90
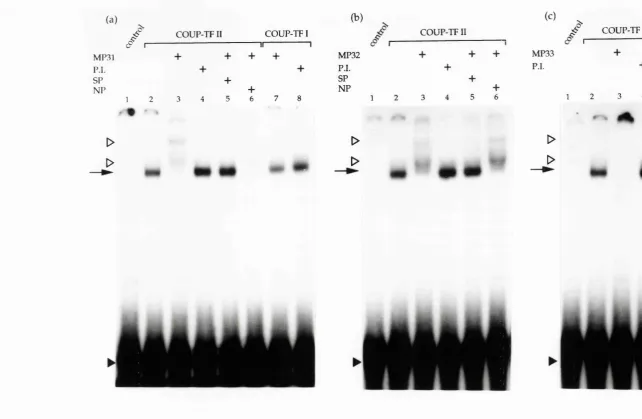
Gel shift analysis of the antisera MP31, MP32 and MP33... 93
Expression of COUP-TF protein in cancer cell lin e s...93
Expression of COUP-TF protein in breast and e n d o m e tria l... tu m o u r sa m p le s... 96
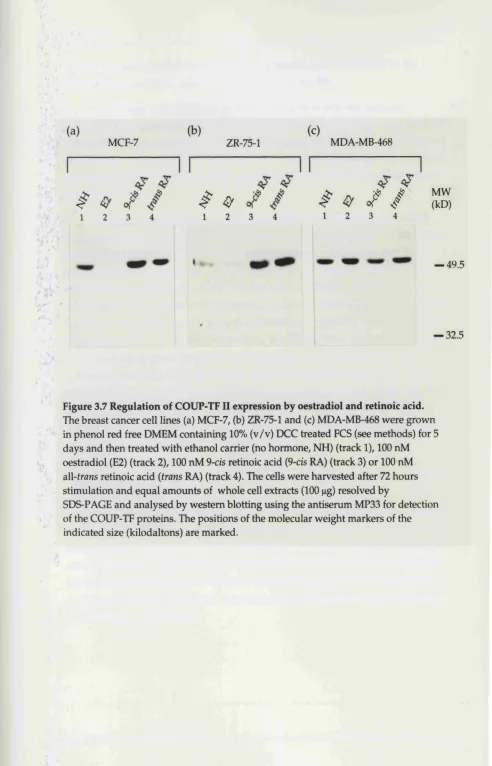
Regulation of COUP-TF expression by oestradiol and retinoic a c id ... 96
Time course analysis of the oestradiol induced dow n regulation of COUP-TF p ro tein ...100
Sum m ary and conclusions...102
C hapter 4 D im érisation properties of COUP-TF II Introduction... 104
COUP-TF II form s both hom odim ers and heterodim ers in v itro... 104
DR+1 response elem ent... 104
DR+2, DR+4 and DR+5 response elem ents...107
T R E pal... I l l SF-1 response e lem en t... I l l Exam ination of the form ation of COUP-TF II hom odim ers and ... h e te ro d im e rs in v iv o...114
Two hybrid a ssa y ...114
Transient transfection assay... 115
R X R a/R A R a but not RXRa/COUP-TF I I ...
heterodim ers can be detected in v iv o...121
R X R a/TR p b u t not COUP-TF II/TRp heterodim ers ... are form ed efficiently in v iv o...125
The deletion m utant CII(DE) does not in h ib it... transcriptional activation by GAL4-TR(DE)...128
The deletion m u tan t CII(DE) does not inhibit 9-cis retinoic a c i d ... stim ulated transactivation from the ApoAI-RARE... 128
COUP-TF II does not recruit VP16RXR to an ApoAI-RARE or a DR+5 response elem ent... 131
Silencing of transcription by COUP-TF II...135
Repression of basal level transcription by GAL4-CII(DE)... 135
Effect of COUP-TF II on basal transcription...137
Sum m ary and conclusions...141
C hapter 5 DNA b in d in g properties of COUP-TF II Introduction... 144
COUP-TF II hom odim ers can bind to response elements ... differing in the spacing and orientation of the h a l f ... site sequence AGGTCA... 144
Exam ination of the binding of COUP-TF II hom odim ers to ... response elements that contain non consensus ... (A/G)GGTCA half sites... 149
A nalysis of the form ation of COUP-TF II hom odim ers on the ... DR+1 and DR+5 response elem en ts...150
The in vitro synthesis of COUP-TF II deletion m u tan ts... 153
Identification of tw o dim érisation interfaces w ithin COUP-TF II...153
Exam ination of the cooperative binding of COUP-TF II on different AGGTC A response elem ents...161
Exam ination of the cooperative binding of COUP-TF II o n ... natural RAREs... 164
Identification of a region adjacent to the zinc finger m otif o f ... COUP-TF II that participates in DNA binding...165
Introduction...169
Effect of ligand on the form ation of R X Ra/TRp heterodim ers in intact cells... 169
Analysis of the effect of ligand on the form ation of RX R a/TR p h etero d im ers in v itr o... 173
Sum m ary and conclusions... 175
Chapter 7 Discussion Introduction...177
Expression of COUP-TF...177
Transcriptional repression by nuclear receptors...179
Form ation of inactive h etero d im ers...179
Com petition for DNA binding...184
S q u elch in g ...186
Repression of other transcription factors...187
R epression of basal transcription- transcriptional silencing...188
COUP-TF as a transcriptional activator...190
DNA binding properties of COUP-TF hom odim ers...193
The role of ligand in dim er form ation...198
A ppendix... 201
Bibliography... 237
List of Figures Chapter 1 1.1 N uclear receptor response elem ents...25
C hapter 3
3.1 C onstruction of a series of 5' and 3' deletion m utants o f ...
COUP-TF I I ... 86
3.2 Im m unoprécipitation of COUP-TF II using the antisera ... MP31, MP32 and MP33...89
3.3 W estern blot analysis of the antisera MP31, MP32 and M P33...92
3.4 Gel shift analysis of antisera MP31, MP32 and M P33...95
3.5 Expression of COUP-TF protein in cancer cell lines...97
3.6 Expression of COUP-TF protein in breast and endom etrial ... tu m o u r sam p les... 98
3.7 Regulation of COUP-TF II expression by oestradiol and ... retinoic acid ... 99
3.8 Time course of the dow n regulation of COUP-TF induced ... by oestradiol...101
C hapter 4 4.1 Models for the mechanism by which COUP-TF represses ... transactivation m ediated by nuclear receptors...105
4.2 D eterm ination of the relative am ounts of the nuclear ... receptor proteins synthesised in vitro...106
4.3 Form ation of COUP-TF II hom odim ers and h e te ro d im e rs... on the DR+1 response elem ent in v itro... 109
4.4 Form ation of COUP-TF II hom odim ers and h e te ro d im e rs ... on the DR+2, DR+4 and DR+5 response elements ... 110
4.5 DNA binding of COUP-TF II and TRp on the DR+4 a n d ... TREpal response elem ents in v itr o...112
4.7 DNA binding of COUP-TF II and SF-1 to the -210 elem ent ... of the steroid 21-hydroxlase gene in v itro... 113
4.7 Schematic representation of the two hybrid assay... 116
4.8 Schematic representation of the chimeric receptors used in the two hybrid assay ...117
4.9 DNA binding activity of GAL4 chim eras...118
4.10 Form ation of COUP-TF II hom odim ers in intact cells...120
4.13 Cotransfection of COUP-TF II or CII(DE) enhances ... transactivation m ediated by GAL4-TR(DE)...129 4.14 COUP-TF b u t not the m utant CII(DE) represses 9 - c is...
retinoic acid transactivation through the ApoAI-RARE...130 4.15 RARa b u t not COUP-TF II can recruit VP16RXR on D N A ... 134 4.16 Com parison of the silencing activity of GAL4-CII(DE) a n d ...
GAL4-TR(DE) on the reporters p G E lb C A T ,... pGSElbCAT and pUASx2-TKLuc...136 4.17 Analysis of COUP-TF II transcriptional activity... 140
Chapter 5
5.1 C om petition for COUP-TF II binding by oligonucleotides... containing variations in the spacing and orientation ... of the AGGTC A half site ...148 5.2 C om petition for COUP-TF II binding by the natural ...
response elem ents ApoAI-RARE and vitellogenin A2 ERE... 151 5.3 DNA binding activity of COUP-TF II hom odim ers to ...
response elements that contain non consensus ... (A/G)GGTCA) half site s...152 5.4 DNA binding activity of COUP-TF II C-term inal deletion m u tan ts 157 5.5 Cooperative DNA binding by COUP-TF II and the deletion
m utant CII-190...158 5.6 DNA binding activity of the deletion m utants CII-155 a n d ...
Cn-144...160 5.7 Sum m ary of the DNA binding properties of the C -...
term inal COUP-TF II deletion m u ta n ts... 162 5.8 DNA binding activity of the deletion m utants CII-155 a n d ...
CII-144 on oligonucleotides containing variations in ... the spacing and orientation of the AGGTCA half site... 163 5.9 Analysis of the DNA binding activity of the deletion ...
Chapter 6
6.1 Effect of T3 on recruitm ent of VP16RXR by CAL4-TR(DE)... and GAL4-TRL454A(DE)...170 6.2 The effect of ligand on the recruitm ent of VP16RXR(DE) by ...
GAL4-TRL454A(DE)...172 6.3 The effect of ligand on the formation of R X R a /T R p ...
heterodim ers on the DR+4 response e lem en t...174
Chapter 7
7.1 Sequence alignm ent of the T box region of m em bers of t h e ... nuclear receptor family...197
List of Tables
Table 1.1 The nuclear receptor superfam ily... 21 Table 5.1 Sequences of the oligonucleotide pairs used in gel shift
A F1/A F2 activation function 1/activ atio n function 2 ARP-1 apolipoprotein AI regulatory protein 1 ATP adenosine 5' triphosphate
A R androgen receptor
BES N, N -bis[2-hydroxyethyl]-2-am inoethanesulphonic acid
bp base pair
BSA bovine serum album in
CAT chloram phenicol acetyl transferase cD N A com plem entary DNA
CEF chicken embryo fibroblast
COUP-TF chicken ovalbum in upstream prom oter transcription factor
C -term in al carboxyl term inal
CTD carboxyl term inal dom ain CTP cytidine 5'-triphosphate
CM V cytom egalovirus
dA TP 2'-deoxyadenosine-5'-triphosphate dCTP 2'-deoxycytidine-5'-triphosphate dCTP 2'-deoxyguanosine-5'-triphosphate dTTP 2'-deoxythym idine-5'-triphosphate DCC dextran coated charcoal
DEAE d ieth y lam in o eth y lam in e
DMEM Dulbecco’s m odified Eagle's m edium DMSG dim ethyl sulphoxide
D N A deoxyribonucleic acid D N ase deoxyribonuclease
DR+X direct repeat w ith X nucleotide spacing DTT d ith io th reito l
EDTA ethylenediam inetetracetic acid
ER oestrogen receptor
ER+X everted repeat w ith X nucleotide spacing ERE oestrogen response elem ent
FCS foetal calf serum
Abbreviations
GRE glucocorticoid response elem ent GST glutathione-S-transferase
GXP guanosine 5'-triphosphate
HEPES N -2-hydroxyethylpiperazine N'-2-ethansulphonic acid
h sp heat shock protein
HRE horm one response elem ent
IR+X inverted repeat w ith X nucleotide spacing
Kb kilobase
kD k ilo d alto n
Kd dissociation constant lacZ p-galactosidase gene
M M TV m ouse m am m ary tum or virus M R m ineralocorticoid receptor m R N A m essenger RNA
NBRE NGFI-B response elem ent
NGFI-B nerve grow th factor inducible protein-B N M R nuclear m agnetic resonance
NP40 nonidet p40
N -te rm in a l am ino term inal
OOx optical density at wavelength of x nm PAGE polyacrylam ide gel electrophoresis PBSA phosphate buffered saline A
PGR polym erase chain reaction
PMSF phenylm ethylsulphonyl fluoride
PPAR peroxisom e proliferator activated receptor
PR progesterone receptor
R A R retinoic acid receptor
RARE retinoic acid response elem ent R N A ribonucleic acid
R N ase ribonuclease
RXR retinoid X receptor
S Svedberg units
SDS sodium dodecyl sulphate SF-1 steriodogenic factor-1 SV40 sim ian virus 40
TBP TATA binding protein
TEMED N 'N 'N 'N '-te tra m e th y le th y le n e d ia m in e
TK thym idine kinase
TR thyroid horm one receptor
TRE thyroid horm one response elem ent
TREpal palindrom ic thyroid horm one response elem ent Tris tris(hydroxym ethyl)am inoethane
Triton X-100 octyl phenoxy polyethoxyethanol Tween-20 polyoxyethylenesorbitan m onoluarate UA S upstream activating sequence
UTP u rid in e 5'-triphosphate
U V u ltra violet
In tro d u ctio n
The cellular processes required for the grow th, developm ent and hom eostasis of an organism involve highly complex patterns of gene expression. A m ultitude of m echanism s are involved in p roviding the required spatial and tem poral regulation and one layer of this control is provided by the action of transcription factors which bind to specific cis acting sequences and m odulate gene transcription. This thesis describes a study of the chicken ovalbum in upstream prom oter transcription factor (COUP-TF), which belongs to a family of transcription factors know n as nuclear receptors. Initial studies on this family focused on the properties of the horm one receptors. More recently, related proteins that have no
identified ligand, of w hich COUP-TF is one example, have also been
investigated. The aim of this chapter is to review our current understanding of the m olecular m echanism s by w hich nuclear receptors function w ithin the cell.
The identification of intracellular horm one receptors
H orm one receptors m ediate the action of m any im portant cell signalling molecules that include the steroid horm ones, thyroid horm one, retinoids and vitam in D. These regulatory molecules play im portant roles in grow th, developm ent and hom oeostasis. The sex steroid horm ones,
androgens, progestins and oestrogen are required for the norm al
developm ent and function of reproductive system s in m am m als (King 1974, C unha et al 1991). The adrenal steroid horm ones, glucocorticoids and
m ineralocorticoids, are involved in the m aintenance of hom eostasis in particular carbohydrate and m ineral m etabolism respectively (Jensen 1991). Vitam in D is required for proper bone form ation and calcium m etabolism (H aussier et al 1988) and the retinoids and thyroid horm ones are involved in grow th and developm ent in m am m als and in addition,
m etam orphogenesis in am phibians (Chin 1991, Kastner et al 1994, Ragsdale and Brockes 1991).
These horm ones can pass readily into the cell and act via intracellular receptors in contrast, peptide horm ones do not enter the cell and their
Chapter 1 Introduction
treated w ith labelled oestradiol, dem onstrated that there w ere two sites of high affinity horm one binding. Most of the oestradiol b ound to the nucleus and a lesser am ount was retained in the cytoplasm. Prior to horm one
treatm ent m ost of the receptor was found in the m ore readily extracted cytoplasmic form. This cytoplasmic receptor complex had a sedm inentation coefficient of 9.5 S while the receptor complex extracted from nuclei
sedim ented at 5 S (Jensen et al 1968, Toft and Gorski 1966). A 'two step' m odel was proposed to account for these observations in w hich u pon b inding horm one the cytoplasmic 9.5 S form of the oestrogen receptor (ER) w as translocated to the nucleus and transform ed into the 5 S form (Jensen et al 1968). The conversion of receptor from a 8-10 S to a 4 S form upon ligand binding has since been dem onstrated for the progesterone, androgen and glucocorticoid receptors (PR, AR and GR respectively) (reviewed in Jensen 1991). This ligand dependent transform ation process could be mimicked in vitro by heat treatm ent and inhibited by some m etal ions such as m olybdate (D ahm er et al 1984).
The ligand induced alteration in the receptor complex involves the dissociation of associated proteins and also seems to involve receptor dim érisation (DeMarzo et al 1991, Miller et al 1985). A num ber of proteins have been identified associated w ith steroid horm one receptors in cell extracts including the heat shock proteins (hsp) 70 and 90, p23, p50, p54, p59 and p60 (for review see Smith and Toft 1993). Hsp 90 is the best characterised of these proteins and dissociates from the receptor upon ligand binding or heat treatm ent (reviewed in Pratt 1993). The ligand sensitive interaction betw een the GR and hsp 90 has also been dem onstrated to occur in intact cells suggesting that the interaction observed in vitro is not an artefact generated during the extraction of the receptor. In pulse chase experim ents labelled hsp 90 could be coim m unoprecipitated w ith the glucocorticoid receptor from untreated cells b u t not from cells treated w ith the synthetic glucocorticoid dexam ethasone (How ard and D istelhorst 1988). In addition, unliganded GR could be crosslinked to hsp 90 in intact cells (Rexin et al 1988). The horm one binding dom ains of the GR and PR are sufficient for the interaction w ith hsp 90, however, binding to the ER requires other regions of the receptor in addition to the horm one binding dom ain (reviewed in Pratt 1993).
expressed in a m utant yeast strain, expressing reduced levels of hsp 90, exhibited reduced transcriptional activity (Picard et al 1990a). H sp 90 m ay act as a chaperone in protein folding, prom oting a structure of the receptor required for receptor function or transport across m em branes (reviewed in Smith and Toft 1993). Hsp 90 also appears to m aintain unliganded steroid horm one receptors in an inactive conform ation although the specific activity it blocks is not clear and m ay differ betw een receptors. It has been suggested that hsp 90 m ight mask the nuclear localisation signal of the GR (Ylikomi et al 1992), however, as this receptor displays a different subcellular distribution to the other steroid receptors it is unlikely that retention in the cytoplasm is a commmon property of hsp 90. A second inhibitory role for hsp 90 was suggested by the report from Sanchez and coworkers (Sanchez et al 1987) that hsp 90 can inhibit the DNA binding activity of the GR in vitro . It is possible that in vivo the DNA binding activity of the steroid horm one receptor is blocked until ligand dissociates the hsp 90 or, alternatively, hsp 90 m ay block protein interactions required for transactivation. The retinoic acid, thyroid horm one and vitam in D receptors (RAR, TR and VDR respectively) do not form stable complexes w ith hsp 90 and, in contrast to the steroid horm one receptors, are 'tightly' bound to the nucleus in the absence of horm one (Dalm an et al 1990, Dalman et al 1991). It rem ains to be determ ined w hether hsp 90 interacts w ith any of the orphan receptors. N uclear localisation
On the basis of cell fractionation experim ents the original tw o step m odel proposed that in the absence of ligand the steroid horm one receptors w ere retained in the cytoplasm. Using im m unocytochem istry, how ever, the ER (King and Greene 1984, W elshons et al 1985), PR (Gasc et al 1989, Perrot- A p p lan at et al 1985) and GR (Govindan 1980, Papam ichail et al 1980, Picard and Yamamoto 1987, W ickstrom et al 1987) have been dem onstrated to enter the nucleus in the absence of horm one. The ER and PR are
Chapter 1 * Introduction
and a horm one inducible NLS is also present w ithin the receptor (Guiochon- M antel et al 1989, Picard et al 1990b, Picard and Yamamoto 1987, Ylikomi et al 1992). The constitutive NLS is sufficient for nuclear targeting of the ER and PR w hereas in the GR this NLS is m asked by the horm one binding dom ain and ligand is therefore required both for the horm one inducible NLS and for revealing the 'constitutive' NLS (Picard et al 1990b, Ylikomi et al 1992). This difference m ay explain the altered cellular localisation of the unliganded GR as com pared w ith the ER and PR.
T he nuclear receptor superfam ily
The first horm one receptors for which com plem entary DNAs
(cDNAs) were obtained were the receptors for glucocorticoid and oestrogen. C om parison of the predicted protein sequence of these receptors identified regions of sequence hom ology suggesting that they m ight belong to a family of ligand activated transcription factors (Green et al 1986, H ollenberg et al 1985). This was confirmed w ith the isolation of cDNA clones for the receptors of the other major steroid horm ones and thyroid horm one, retinoic acid, vitam in D (reviewed in Beato 1989, Evans 1988, G reen and C ham bon 1988, H am and Parker 1989, Parker 1993) and m ost recently for farnesol (Form an et al 1995). The clones for the TR w ere show n to be the cellular hom ologues of the viral oncogene v -e rb A found in the avian erythroblastosis virus (Sap et al 1986, W einberger et al 1986). O ther
m em bers have been isolated th at do not have an identified ligand, the so- called orphan receptors. To date there are approxim ately 30 different m em bers of this family identified in vertebrates, two thirds of w hich are o rphan receptors (Table 1.1) and a num ber of nuclear receptor hom ologues have also been identified in D rosophila m elanogaster (Table 1.1) (review ed in O ro et al 1992, Segraves 1994).
Construction of phylogentic trees illustrates that the nuclear receptor fam ily evolved from a single progenitor protein (Amero et al 1992, L audet et al 1992). The only exceptions being the three Drosophila proteins kn irp s, knirps-related, and em bryonic gonad that have acquired a different C- term inal dom ain d uring evolution. Analysis of genomic clones for several nuclear receptors revealed a sim ilar e x o n /in tro n arrangem ent also
suggesting that these receptors originated from a comm on precursor. In general, the N -term inus is encoded by a single exon and is seperate from the DNA binding dom ain which is encoded by two exons, w ith the intron
COUP-Glucocorticoid GR
Progesterone PR
M ineralocorticoid M R A R ER A ndrogen
O estrogen
Retinoic acid Retinoid X
T hyroid horm one Vitam in D
Farnesoid X
RAR a /p /y RXRa/p/y TRa/p VD R FXR
COUP-TF I (Ear-3)/COUP-TF II (ARPl) Ear-2
HNF-4 Tlx (til) TR2 TR4 ERR 1 /2 LXR
PPA R a/p/y RLDl MB67 RIP 15
Rev-erba (Ear-l)/R ev-erbp (BD73/RVR) R Z R /R O R a/p
NGFI-B (N urr 77) N U R R l
RNR-1 LRHI
SF-1 (Ad4BP/ELP) GCNF
Seven-up (svp)
HNF-4(D)
Tailless (til)
U ltraspiracle (usp) Ecdysone Receptor (EcR)
E75 E78 DHR3 DHR39
Fushi-tarazu F la /p (FTZ-Fla/p)
K nirps* (kni)
K nirps-related* (knrl) Embryonic gonad* (egon) Table 1.1 The Nuclear Receptor Superfam ily
Chapter 1 Introduction
TF lacks the intron betw een the two zinc fingers (Ritchie et al 1990) and another orphan receptor, NGFI-B, lacks the intron betw een the N -term inus and the first zinc finger (Ryseck et al 1989).
A num ber of receptors, including the horm one receptors RAR, TR and retinoid X receptor (RXR), are encoded by m ultiple genes. The
complexity is increased further by the observation that different isoforms of the gene products can be produced by alternative prom oter usage and
differential splicing (reviewed in Chin 1991, Leid et al 1992a). The receptors for retinoic acid display the greatest diversity w ith two m ain types of
receptor, the RARs which bind both 9-cis and all-trans retinoic acid and the RXRs which bind 9-cis retinoic acid only. Both of these receptors are encoded by m ultiple genes (a, p, y) and different isoforms of these receptors are also observed (see Leid et al 1992a). Some orphan receptors also appear to have undergone gene duplication, for example COUP-TF has two identified loci (I and II) (Ritchie et al 1990). In general, a specific receptor isoform shows higher sequence conservation betw een species than betw een different receptor isoforms from the same species.
COUP-TF
COUP-TF I and II are highly homologous, with the DNA binding domains differing by a single conservative amino acid change (threonine/ serine) and the C-terminal region sharing 97% amino acid identity. Within the nuclear receptor family the COUP-TFs are most highly related to the orphan receptor ear-2 (Miyajima et al 1988). Interestingly, COUP-TF also shares regions of strong homology with RXR, indeed RXR is more highly related to COUP-TF than to RAR (Leid et al 1992a).
The gene seven-up is the Drosophila homologue of COUP-TF
(Mlodzik et al 1990) and the sequence conservation between these proteins is particularly high, with greater than 85 %^identity between the DNA binding domains and the C-termini. The overall conservation between the two proteins is striking (>75%) and is higher than for any other known set of Drosophila and vertebrate homologues (Fjose et al 1993). COUP-TF homologues have also been identified in Xenopus laevis, zebrafish
{Brachydanio rerio) and sea urchin {Strongylocentrotus purpuratus) (Chan et al 1992, Fjose et al 1993, Matharu and Sweeney 1992) demonstrating that this protein has been highly conserved during evolution. In Drosophila, seven- up is involved in the determination of photoreceptor neurons during eye development. A role for COUP-TF in early neurogenesis is supported by the predom inant expression of the zebrafish homologues [44] and [46] in the developing central nervous system and eye (Fjose et al 1993). Furthermore, m urine COUP-TF homologues are expressed at high levels in the central nervous system during development (Qiu et al 1994) and a chicken COUP- TF homologue is expressed in developing motor neurons and brain (Lutz et al 1994). At present the mechanism of COUP-TF function during
developm ent is unclear although in Drosophila it appears to require an active ras signalling pathway (Begemann et al 1995). In transfected cells COUP-TFs can repress the activity of other nuclear receptors including RXR. Intriguingly the Drosophila protein usp, which is the homologue ^
vertebrate RXR, is also required for normal eye development and it is tem pting to predict that seven-up restricts the activity of this protein. O ther orphan receptors
Chapter 1 Introduction
Sladek et al 1990). NGFI-B was cloned by several groups as an early response gene w hich is rapidly and transiently induced by grow th factors such as nerve grow th factor (Hazel et al 1988, W atson and M ilbrandt 1989) and
another related im m eadiate early gene, RNRl, has since been cloned (Scearce et al 1993). A protein interaction screen in yeast identified RIP 15 as an
o rphan receptor that interacts w ith the ligand binding dom ain of RXR (Seol et al 1995) and the gene for Rev-erba (ear-1) was discovered by virtue of it being encoded on the noncoding strand of the TRa gene (Lazar et al 1989b). N uclear receptor response elem ents
M embers of the nuclear receptors family can be divided into three general classes on the basis of their DNA binding activity. Class I contains the steroid horm one receptors which bind as hom odim ers to palindrom ic
sequences (Figure 1.1). Steroid response elements were initially identified th ro u g h deletion analysis of target gene prom oters and alignm ent of glucocorticoid response elements (GREs) and oestrogen response elements (EREs) revealed that they contained sim ilar palindrom ic sequences. T he replacem ent of just two base pairs in the palindrom e converted the ERE of the X enopus vitellogenin A2 gene into a GRE (Klock et al 1987, M artinez et al 1987). From point m utagenesis the nucleotides required for receptor recognition were discrim inated and consensus GRE and ERE sequences identified (Figure 1.1) (see M artinez and Wahli 1991). The sym m etry of the sites suggested that these receptors bound to DNA as dim ers and this was confirm ed using gel shift analysis (Kumar and C ham bon 1988). Surprisingly, the PR, AR and the mineralocorticoid receptor (MR) could also stim ulate transcription from a GRE (Cato et al 1986, Chalepakis et al 1988, Darbre et al 1986, H am et al 1988, Strahle et al 1987). A lthough these receptors bind com m on response elements it seems that their DNA binding dom ains m ake different contacts, producing distinct DNase I footprinting and m éthylation interference patterns (Chalepakis et al 1988, von et al 1985). The receptors also displayed different sensitivites to alterations in the response elem ent sequence (Cato et al 1988, Chalepakis et al 1988).
The class II receptors bind to response elements com posed of direct, inverted and everted repeats of a hexam er sequence sim ilar to that of an ERE half site (see Figure 1.1). This group of receptors includes the TR, RAR, VDR and RXR as well as m any of the orphan receptors. M utagenesis of the thyroid horm one repsonse elem ent (TRE) from the rat grow th horm one gene
Class I
AGRACANNNTGTMCY GRE(IR+3) GR, PR, MR, AR
AGGTCANNNTGACCT ERE (IR+3) ER
Class II
RGGTCANxRGGTCA DR+X
RGGTCANxTGACCY IR+X
TGACCYNxRGGTCA ►
ER+X
Class III
RXR, RAR, TR, VDR, COUP-TF, PPAR,
AAAGGTCA NBRE NGFI-B, NURRl
YCAAGGTCA FRE FTZ-Fl, SF-1, ELP
WAWNTAGGTCA ►
RevRE RZR/ROR,
Rev-erb/RVR
Figure 1.1 N uclear receptor response elem ents.
Chapter 1 Introduction
arrangem ent of the sequence AGGTCA and is referred to as a TREpal (Brent et al 1989, Glass et al 1988). This element w as similar to the consensus ERE w ith the exception that the three base pairs separating the repeats were m issing in the TREpal. This element was also show n to be retinoic acid inducible (Glass et al 1989, Umesono et al 1988) and subsequent reports have sim ilarly dem onstrated that the response elem ent specifcity of the thyroid horm one and retinoic acid receptors can overlap (G raupner et al 1989, M angelsdorf et al 1990).
The observation that the TRE identified in the rat m yosin heavy chain prom oter conferred T3 b u t not retinoic acid inducibility led to the discovery of another class of binding site. The AGGTCA related half sites of this
elem ent are arranged in a direct repeat arrangem ent w ith a four nucleotide spacing betw een the repeats. The use of natural and idealised sites led to the observation that direct repeats w ith different spacings w ere selective for different receptors (Naar et al 1991, Um esono et al 1991) The VDR bound preferentially to a direct repeat w ith a three base pair spacing (DR+3), TR to a four (DR+4) and RAR to a five nucleotide spacing (DR+5). This selectivity w as also observed in transient transfection experim ents w ith a DR+3
From in vitro DNA binding studies it w as apparent th at a cofactor p resent in nuclear extracts enhanced the binding activity of RAR, TR and VDR (Burnside et al 1990, Glass et al 1990, M urray and Towle 1989). These factor(s) form ed novel heterodim eric complexes w ith RAR or TR that had higher binding affinity than the hom odim eric receptors (Glass et al 1990, Lazar et al 1991, N aar et al 1991). Intriguingly, the TR had been show n to form a heterodim er w ith RAR in vitro that bound to various TREs w ith ten fold greater affinity than the TR hom odim er (Glass et al 1989) although as this heterodim er did not bind w ith increased affinity to an RARE it was unlikely that TR w as the RAR cofactor present in nuclear extracts. This observation did suggest, however, that the stim ulatory factor m ight be
another nuclear receptor and conform ation came w hen the RAR coregulator w as isolated and show n to be identical to RXR (Leid et al 1992b, Marks et al 1992, Y u et al 1991).
RXRa was originally cloned by M angelsdorf et al (M angelsdorf et al 1990) by virtue of its hom ology to RARa and H am ada et al (H am ada et al 1989) had cloned RXRp, also know n as H-2RIIBP, by its interaction w ith region II of the major histocom patibility complex class I genes. A lthough originally classified as an orphan receptor it has since been show n to be b ound and activated by 9-cis retinoic acid (Heyman et al 1992, Levin et al 1992). RXR enhances the binding of RAR, TR and VDR to inverted and direct repeats response elements (Kliewer et al 1992b, Leid et al 1992b, Marks et al 1992, Yu et al 1991, Zhang et al 1992a) and similarly increases the DNA binding activities of some orphan receptors including PPAR, LXR, RLD-1 and MB67 (Apfel et al 1994, Baes et al 1994, Gearing et al 1993, Kliewer et al 1992c, Willy et al 1995). The majority of the receptors of the class II group tested so far bind preferentially as a heterodim er to direct and inverted repeat response elements. The form ation of heterodim ers has been conserved d u rin g evolution as the DNA binding and transactivation of the D rosophila ecdysone receptor is enhanced through the form ation of heterodim ers w ith ultraspiracle the D rosophila hom ologue of RXR (Thomas et al 1993, Yao et al 1992). Strikingly, U ltraspiracle can substitute for RXR in form ing
heterodim ers w ith RAR, TR, VDR and PPAR w hile RXR can form heterodim ers w ith the ecdysone receptor (Yao et al 1992).
A lthough TR hom odim ers only bind w eakly to direct and inverted repeats they can bind w ith high affinity to everted repeats spaced by 4, 5 or 6 base pairs (Kurokawa et al 1993, N aar et al 1991, Piedrafita et al 1995).
yF-Chapter 1 Introduction
crystallin gene, composed of an everted repeat w ith an 8 base pair spacing, is b o u n d preferentially by RXR/RAR or TR/RA R heterodim ers (Tini et al 1994). A TR/VDR heterodim er has also been detected on everted repeats w ith 7 and 9 base pairs (Schrader et al 1994b).
Response element selectivity for the class II group of receptors is therefore rather complex and is dictated by a variety of factors including w hether the receptor binds as a hom odim er or heterodim er, the orientation, spacing and sequence of the half sites and in addition, the sequence of the flanking nucleotides. W hen the w ork described in this thesis w as initiated COUP-TF had only been show n to bind DNA as a hom odim er. H ow ever, d u rin g the course of this thesis it become apparent that COUP-TF m ay also form heterodim ers and this question is addressed in the w ork described in C hapter 4.
The third class of receptors consists of orphan receptors that bind DNA as m onom ers and includes NGFI-B, SF-1, Rev-erb and the Drosophila F T Z- F l. W hen the group of M ilbrandt and coworkers (Wilson et al 1991)
originally identified a response elem ent for NGFI-B they m ade the
surprising discovery that it contained a single half site motif. The failure to detect a heterodim er betw een w ild type and truncated protein on this site in gel shift analysis supported the prediction that NGFI-B bound as a m onom er (cited in W ilson et al 1992). M utational analysis of the elem ent identified nucleotides im portant for receptor bindng in vitro and the sequence AAAGGTCA was proposed as a consensus NGFI-B response elem ent (NBRE) (Wilson et al 1992). The other m em bers of this group also bin d as m onom ers to response elements that consist of an extended half site m otif w ith the nucleotides im m eadiately 5' of the half site involved in specifying receptor selectivity (Figure 1.1) (Ueda et al 1992, W ilson et al 1993). In
addition, RZR (also know n as ROR) can also bind as a hom odim er to certain sites that contain two half site motifs (Carlberg et al 1994). More recently, NGFI-B and NU RRl have been reported to form heterodim ers w ith RXR (Form an et al 1995, Perlm ann and Jansson 1995) and it is possible that receptors from this class m ay bind DNA either as a m onom er, hom odim er or heterodim er depending on the type of response element.
transcription factors as well (Carter et al 1994, Chalepakis et al 1988, M artinez et al 1987, reviewed in M artinez and Wahli 1991). On such sites synergistic transcriptional activation can be observed. This occurs w hen the
transcriptional activation due to two or m ore transcription factors is greater than the sum of the activation observed w hen each is bo u n d individually (Ptashne 1988) and this synergy is due to cooperation of the factors in binding DNA or stim ulating transcription (reviewed in M artinez and W ahli 1991). N uclear receptor structure
On the basis of sequence similarity betw een the predicted protein sequences of the ER and GR several regions (A-F) were defined (Krust et al 1986). These regions display different degrees of evolutionary conservation am ongst the m em bers of the nuclear receptor family. Region C, the DNA binding dom ain, is highly conserved betw een the different receptor types w hereas, in contrast, the N-term inal regions A /B are highly variable both in sequence and size, varying from 24 amino acids in the VDR to 603 in the (MR). Similarly, region D shows poor sequence conservation betw een receptor types. Region E is the largest dom ain and is involved in ligand binding, dim érisation and transactivation. This region is only m oderately well conserved presum ably reflecting the different ligand specificties of the receptors. Region F, the sequence C-terminal to the ligand binding dom ain, is poorly conserved and is lacking in some receptors altogether. Between different species each receptor type is well conserved w ith regions C and E containing the highest homology. In particular, region C show s over 90% co nservation.
The m odular structure of nuclear receptors has been a significant advantage in the analysis of their functional properties. The aim of the following section is to provide an overview of the DNA binding, dim érisation, ligand binding and transactivation properties of nuclear receptors.
DNA b in d in g
The first experim ental evidence that region C contained the DNA b inding dom ain came from 'dom ain sw ap' experim ents. G reen and
Chapter 1 Introduction
transcription factor IIIA (TFIIIA). In the TFIIIA motif each zinc ion w as tetrahedrally coordinate by two cysteines and two hystidines (Miller et al 1985) and by analogy it was suggested that region C contained tw o zinc finger DNA binding motifs, w ith the zinc ions tetrahedrally coordinated by cysteine residues. Evidence supporting the existence of a zinc finger DNA binding m otif w as provided by m utagenesis of the GR. Significantly, w hilst
replacem ent of eight of the conserved cysteine residues of region C to alanine or serine residues abolished DNA binding, replacem ent w ith histidines retained the activity (Severne et al 1988). Furtherm ore, m easurem ents m ade using extended X-ray absorption fine structure
suggested that the DNA binding dom ain contained two zinc ions w ith each ion coordinated by four cysteines (Freedman et al 1988).
A dditional m utagenesis studies of the ER and GR identified
individual am ino acid residues involved in providing the response elem ent specificty. Green et al (Green and Cham bon 1988) initially localised this p ro p erty to the N -term inal zinc finger. The replacem ent of tw o am ino acids, betw een the third and fourth cysteine of the N-term inal zinc finger of the GR, w ith the corresponding ER residues resulted in a m utant that stim ulated transcription efficiently from an ERE and poorly from a GRE (Danielsen et al 1989, Um esono and Evans 1989). To convert the ER to specifically recognise a GRE a residue located betw een the zinc fingers was required in addition to the tw o amino acid residues identified in the GR (M ader et al 1989). These three amino acids are referred to as the P box. In the ER the P box residues are glutam ine, glycine and alanine w hile in the GR they are glycine, serine and valine. The GR P box sequence is conserved betw een the PR, MR and AR, w hich all recognise a GRE. Those receptors that recognise the sam e half site sequence as the ER contain the glutam ine and glycine residues of the P box b u t not the third residue, alanine. The identity of the third residue is
conserved betw een receptors of the same sub group, for instance, COUP-TF, seven -u p and ear-2 contain a serine at the third position and TR, RAR, RXR and VDR contain a glycine. In contrast, only the first residue of the P box of SF-1 and FTZ-FI is conserved w ith the ER, the second and th ird residues being replaced by serine and glycine.
recognising the particular spacing and are required for the form ation of the dim er interface on their respective response elem ents (Hirst et al 1992, M ader et al 1993a, Umesono and Evans 1989).
NMR analysis of the 3D structure of the DNA binding dom ains of the ER and GR in solution and X-ray crystallographic analysis of their structure on DNA confirm ed and extended the results obtained from the m utagenesis studies. In solution the DNA binding dom ains are m onom eric and the two zinc finger motifs are folded to form a single dom ain, unlike the zinc finger motifs of TFIIIA w hich form independent units (H ard et al 1990b, Schwabe et al 1990). The DNA binding dom ains contain two am phipathic a helices, that start at the C-terminal pair of cysteines of each zinc finger and are arranged at right angles relative to each other. The region betw een the two fingers is poorly ordered w ith a num ber of hydrophobic side chains form ing a hydrophobic core betw een the helices. Resolving of the crystal structure of these receptors on DNA confirmed that two molecules of the DNA binding dom ain bind cooperatively to adjacent major grooves on the same side of the double helix. As predicted, the N-term inal zinc finger is involved in sequence discrim ination w ith one of the P box residues m aking contact w ith a nucleotide from the half site in both the G R /D N A and E R /D N A
complexes (Luisi et al 1991, Schwabe et al 1993). Recognition of response elem ents by the ER and GR appears to involve not only the use of these discrim atory am ino acids that favour interactions w ith different nucleotides b u t also a global change in the arrangem ent of the side chains of common residues (Schwabe et al 1993).
Residues in the D box form p art of a dim érisation interface betw een the DNA binding dom ains (Luisi et al 1991, Schwabe et al 1993). This region in the ER DNA binding dom ain is disordered in solution and the dom ain is m onom eric and it seems that binding of the receptor to DNA is required for the correct interface to be formed (Schwabe et al 1993). W hether the
corresponding region of the GR DNA binding dom ain is disorderd in solution is less clear (Baumann et al 1993, Berglund et al 1992, H ard et al 1990a, H ard et al 1990b, Luisi et al 1991).
Chapter 1 Introduction
the stability of the receptor/D N A complex (Luisi et al 1991, Schwabe et al 1993).
The ten carboxy amino acids of region C, im m eadiately following the second am phipathic a helix, are required for stable binding of the ER on DNA (Schwabe et al 1993). These residues are unstructured in both the NMR and X-ray crystallographic analysis (Schwabe et al 1993, Schwabe et al 1993). M ader et al (M ader et al 1993a) have show n that amino acids in region D are also involved in stabilising DNA binding of the ER especially on imperfect palindrom es. The involvem ent of residues dow nstream of the zinc finger m otifs in high affinity DNA binding w as originally dem onstrated for RXR and the o rphan receptor NGFI-B (Wilson et al 1993) and these additional residues, the T box in RXR and the A box in the TR, have been show n to form a helices (discussed later) (Lee et al 1993, Rastinejad et al 1995). It is possible that the corresponding residues of the ER also form an additional structure that is disordered by crystal packing or, alternatively, the
unstructured residues m ay provide a num ber of weak non specific contacts w ith DNA. Throughout this thesis the conserved zinc finger m otif (residues 180-249 of the hum an ER) (Mader et al 1993a) is defined as the 'core DNA binding dom ain', distinct from the region of the receptor required for high affinity binding.
Reports from several groups indicated that nucleotides flanking the half sites influenced the affinity w ith which receptors bound DNA (Katz and Koenig 1994a, Katz et al 1995, Kurokawa et al 1993, M ader et al 1993; Wilson et al 1992) and this effect w as particularly m arked for those receptors that b ound as m onom ers, for example, NGFI-B, SF-1 and FTZ-Fl (Lala et al 1992, L avorgna et al 1991, W ilson et al 1993, W ilson et al 1992). In binding site selection both RXR/TR and RXR/RAR exhibited partial sequence selectivity of the nucleotides at the 5' of half sites (Kurokawa et al 1993). Supporting evidence for the involvem ent of flanking nucleotides w as provided by DNA m éthylation experim ents w hich indicated that the heterodim ers m adç
m inor groove contacts upstream of the half site motifs (Kurokawa et al 1993). Of interest w as the observation by Wilson et al (Wilson et al 1992) that
heterodim er residues from the A box of the TR w ould form D N A contacts w ith the 5' end of each half site. H igh affinity DNA binding by RXR
hom odim ers to direct repeats w ith a one base pair spacing required a region (the T box) that was distinct, but adjacent to, the A box (Wilson et al 1992).
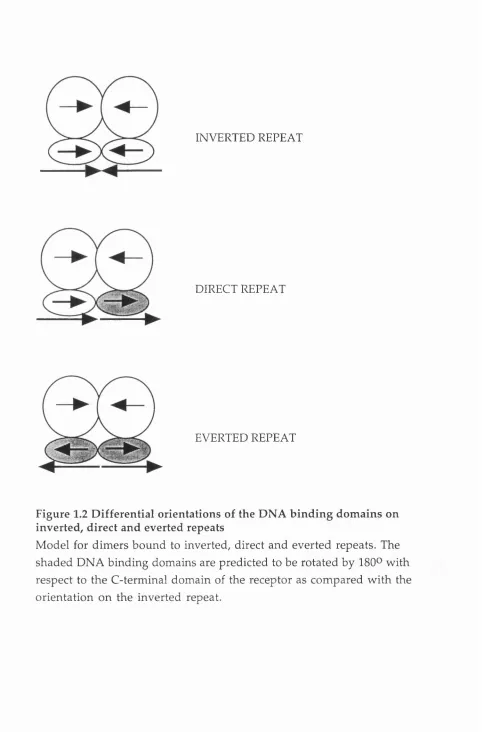
O n direct repeat elements a novel asymmetric interface betw een the DNA binding dom ains of the receptors m ust be predicted as the symmetrical interface used on inverted repeats w ould not be properly aligned (Figure 1.2). Especially intriguing is the ability of the DNA binding dom ain of RXR to bind cooperatively, as a heterodim er, to direct repeats w ith half site spacings of one to five base pairs. NMR structural analysis confirmed that, like the steroid horm one receptors, the two zinc finger motif of RXRa form ed a single dom ain w ith two a helices perpendicular to each other. In contrast to the ER and GR, however, residues C-terminal to the zinc finger m otif
form ed a third a helix (Lee et al 1993). These residues were located in the T box (Wilson et al 1993) and m utagenesis studies indicated that am ino acids w ithin the helix w ere involved in both contacting DNA and in form ing p ro te in /p ro te in contacts betw een m onom ers (Lee et al 1993). C om puter m odelling suggested that the binding of two RXR DNA binding dom ains to a DR+1 repeat w ould prom ote a dim érisation interface betw een the third helix of the RXR m onom er on the 3' half site and the C-terminal zinc finger of the m onom er on the 5'site.
The heterodim ers RXR/TR and RXR/RAR preferentially recognise direct repeat elem ents on which their DNA binding dom ains bind
cooperatively. Studies on these discrim inatory interfaces have im plicated distinct regions w ithin the different receptor types. Perlm ann and coworkers (P erlm ann et al 1993) dem onstrated that on a DR+5 elem ent the cooperative binding of RXR and RAR involved the D box of RXR and the N -term inal zinc finger m otif of RAR and, similarly, the binding of an RXR/TR
heterodim er on a DR+4 elem ent requires the D box of RXR and the N-
INVERTED REPEAT
DIRECT REPEAT
EVERTED REPEAT
Figure 1.2 Differential orientations of the DNA binding dom ains on inverted, direct and everted repeats
on w hich the T box of RAR and a region of the C-terminal zinc finger of RXR distinct from the D box were required. Thus RAR and RXR appear to form distinct interfaces on the different spacings.
The form ation of an asym metric dim érisation interface betw een the DNA binding dom ains of RXR and its heterodim er partners RAR and TR im plied that these heterodim ers bind DNA w ith polarity. W ork from several groups has supported this prediction, an RXR/TR heterodim er on a DR+4 elem ent is orientated w ith RXR on the 5' half site and TR on the 3' half site. Similarly, an RXR/RAR heterodim er on a DR+2 or DR+5 elem ent binds w ith RXR on the 5' half site (Kurokawa et al 1993, M ader et al 1993Ç- P erlm an n et al 1993, Zechel et al 1994b). On a DR+1 element, how ever, the polarity of the RXR/RAR heterodim er is reversed w ith RAR placed on the 5' half site (Kurokawa et al 1994).
The determ ination of the crystal structure of the DNA binding dom ains of a RXR/TR heterodim er bound to a DR+4 response elem ent (Rastinejad et al 1995) has refined the m odels predicted from the
m utagenesis studies. As predicted RXR was located on the 5' half site and the dim er interface, which is formed over the m inor groove of the DNA,
involved different regions of the individual receptors. The C -term inal zinc finger of RXR is involved in the dim er interface, including residues from the D box region w hereas three regions of the TR are involved, the
'prefinger' region of the N-term inal finger, the N -term inal finger and the T box. As observed for the ER and GR, residues in the dim er interface are also involved in contacting DNA illustrating the interplay betw een high affinity DNA binding and dim er formation.
As for the ER and GR, the sequence specificty of the RXR/TR heterodim er is provided by the N-term inal a-helix that m akes specific
Chapter 1 Introduction
m onom er (Katz and Koenig 1994b, Schrader et al 1994a). The o rphan receptors that bind to DNA as m onom ers are predicted to form extensive DNA contacts that obviate the requirem ent for the additional free energy provided by cooperative binding of molecules. By analalogy to the TR it is likely they contain an A-helix type structure that provides som e of the
required p ro tein /D N A contacts (Ueda et al 1992, W ilson et al 1993, W ilson et al 1992).
C om puter m odelling and m utagenesis studies predict tw o m echanism s by which the RXR heterodim ers discrim inate the spacing betw een half sites. The first is that the correct stereochemical alignm ent of the tw o m onom ers required to form a stable interface only occurs on certain spacings. The second is dictated by steric hindrance, for example, RXR/TR heterodim ers are predicted to be blocked from binding direct repeats spaced by less than four base pairs due to the long A-helix of the TR (M ader et al 1993b, Rastinejad et al 1995, Zechel et al 1994b). Steric hindrance is sim ilarly thought to prevent two molecules of ER binding to inverted repeats
seperated by less than three base pairs (Schwabe et al 1993).
The T box of RXR is unstructured in the crystal structure of RXR/TR in contrast to the a-helical structure observed by NMR analysis of the isolated DNA binding dom ain (Lee et al 1993, Rastinejad et al 1995). The T box of the TR, bound to the 3' motif of the DR+4 site forms a loop that is
involved in the RXR/TR dim er interface. It is possible that the T box of RXR is also required on those sites on which it binds to the 3' half site of the elem ent, such as the DR+1 elem ent either as a RXR hom odim er or RAR/RXR heterodim er (Lee et al 1993, Zechel et al 1994b).
D im érisation
As discussed in the previous section the majority of nuclear receptors bind to DNA as dimers. Full length ER forms dim ers in solution as well as on DNA (Fawell et al 1990a, K um ar and Cham bon 1988) w hile in contrast the isolated DNA binding dom ain requires DNA to dim erise and is
et al 1989, Glass et al 1989, Kurokawa et al 1993, Ladias and K arathanasis 1991, Leid et al 1992b, Marks et al 1992, Rosen et al 1993, Selmi and Samuels 1991, Spanjaard et al 1991) and is often referred to as the m ajor dim érisation activity.
Sequence alignm ent of a num ber of receptors identified a conserved hep tad repeat of hydrophobic residues w ithin region E (Fawell et al 1990a). Point m utagenesis of the m ouse ER suggested that residues critical for
dim érisation w ere located in the N -term inal half b u t not the C-term inal half of this m otif (Fawell et al 1990a). Form an et al (Forman et al 1989) identified nine heptad repeats w ithin the equivalent region of the TR (the conserved region identified by Fawell et al (Fawell et al 1990a) overlaps the ninth heptad). These heptad repeats were rem iniscent of the coiled-coil
dim érisation m otif of the leucine zipper and helix-loop-helix transcription factors although structurally distinct and were suggested to form five a- helices that created a 'regulatory zipper'(Form an and Samuels 1990, Form an et al 1989).
H om odim ers of RAR, TR and RXR, unlike heterodim ers, are not readily detected off DNA (Barettino et al 1993, Glass et al 1990, H erm ann et al 1992, Kliewer et al 1992b, Leid et al 1992b, Zhang et al 1992a) im plying that the interface form ed by region E is w eaker in the hom odim er than the
heterodim er and requires DNA for stabilisation. Furtherm ore, the form ation of RXR/TR heterodim ers in solution is favoured over DNA bo u n d TR
hom odim ers (K urokaw a et al 1993) thus, for the TR at least, heterodim eric interactions w ith RXR off DNA seem to be stronger than hom odim eric contacts on DNA. The isolated C-term inal dom ain of RAR forms
hom odim ers as efficiently as heterodim ers w ith RXR (Kurokawa et al 1993) indicating th at the N -term inus or the DNA binding dom ain of RAR m ust restrict the form ation of the hom odim er interface.
M utagenesis studies have revealed that the C-term inal hom odim er and heterodim er interfaces of TR, RAR and RXR are not superim posable. Point m utagenesis identified m utations in the ninth heptad repeat of TR and RAR w hich destabilise heterodim ers w ith RXR b u t do not affect
hom odim er form ation (Au et al 1993). Residues w ithin the second and third heptads of the TR and residues N-term inal to the heptad repeats in RAR and TR have also been im plicated in heterodim erisation w ith RXR (Darling et al 1991, O 'D onnell et al 1991, Rosen et al 1993, Spanjaard et al 1991).
Chapter 1 Introduction
hom odim ers and heterodim ers are distinguishable, C-term inal deletion analysis of RXR suggests that hom odim erisation involves residues C- term inal to those sufficient for heterodim erisation although the ninth h ep tad repeat is required for both the hom odim er and heterodim er functions of RXR (Leng et al 1995, Marks et al 1992, Zhang et al 1994).
The crystal structure of region E of hum an RXRa has recently been solved (Bourguet et al 1995). The dom ain contains eleven a-helices in a three layer structure that folds to form a novel antiparallel a-helical sandwich. This "sandwich" exists as a dim er w ith sym m etrical contacts provided by two of the helices, helix ten and, to a lesser extent, helix nine. An intervening loop structure betw een helix seven and eight also forms p art of the dim er interface. Interestingly, although residues of the ninth heptad are located in helix 10 their side chains are packed aw ay from the dim er interface. Indeed, the hydrophobic residues of all the heptad repeats seem to to be involved in stabilising the a-helical structure rather than being
positioned at the dim er interface. As these hydrophobic residues are conserved it is likely that the overall fold of the dom ain will be com m on throughout the family. Differences in the residues at the dim er interface m ay then specify the dim érisation properties of the receptors, such as w hether it form s hom odim ers exclusively, like the steroid horm one receptors, or can also form heterodim ers.
The DNA binding dom ain of the receptor bound to the 3' half site of a response elem ent is predicted to be rotated by 180® on a direct repeat in com parison to the orientation on a inverted repeat. On an everted repeat both DNA binding dom ains w ould be rotated by 180® (see Figure 1.2). To accom m odate the different orientations of the DNA binding dom ain a flexible hinge region connecting the DNA binding dom ains to the C- term inal dom ain has been proposed. This hinge region corresponds to region D w hich contains the A- and T boxes, residues from these boxes have been suggested to be involved in providing the differential orientations of the DNA binding dom ain (Kurokawa et al 1993).
Ligand B inding
integrity of the entire region, of approxim ately 250 amino acids, appeared to be required for efficient binding suggesting that this region folded as a single dom ain. This was supported by studies that show ed that the horm one binding dom ain was retained in a 28 kD protease resistant fragm ent of the ER and GR (Eisen et al 1985, Katzenellenbogen et al 1987). Region E contains a num ber of conserved hydrophobic residues and it has been proposed to form a hydrophobic 'pocket' in which the ligand binds. Ligand affinity
labelling has identified residues that are part of the ligand binding site and in addition point m utagenesis has identified residues that are required for ligand binding (reviewed in Dauvois and Parker 1993, M cPhaul 1993). Region E also contains the major dim érisation interface (see above), po in t m utagenesis of the m ouse ER (Fawell et al 1990a) and rat TRa (Spanjaard et al 1991) suggests that these two functions overlap b u t are not
superim posable. Analysis of the crystal structure of region E of RXR, determ ined in the absence of ligand, has lead to the identification of a potential ligand binding site. The putative site consisits of a hydrophobic cavity created by four surrounding a-helices into which a molecule of 9-cis retinoic acid can be m odelled w ith little adjustm ent. This m odel fits well w ith the m utagenesis and crosslinking data obtained w ith other nuclear receptors. It is intriguing that although RAR and RXR both bind 9-cis retinoic acid their ligand binding dom ains display only m oderate sequence conservation. This region of m ouse RXRa shares approxim ately 30% am ino acid identity w ith m ouse RARa com pared w ith 44% identity w ith m ouse COUP-TF I (Leid et al 1992a). This divergence m ay have been required to provide RAR, unlike RXR, w ith the ability to bind all-frflMS-retinoic acid. T ranscriptional activation dom ains
Two autonom ous activation functions, AF-1 and AF-2, have been identified in m ost of the horm one receptors w ith the relative contribution of these two activities varying betw een the different receptors (review ed in Danielsen 1991, Green and Cham bon 1991, Gronem eyer 1993). AF-1 is located in the N -term inus (A /B region) and can stim ulate transcription in a ligand independent m anner w hen fused to a heterologous DNA binding dom ain or w hen the C -term inus of the receptor is deleted (Berry et al 1990, G ronem eyer et al 1991, Lees et al 1989, Meyer et al 1990, N agpal et al 1993, Tora et al