organic papers
Acta Cryst.(2007). E63, o1171–o1172 doi:10.1107/S1600536807004813 Liet al. C
34H32N4O2
o1171
Acta Crystallographica Section E Structure Reports
Online
ISSN 1600-5368
c
-2,
t
-4-Bis(2-benzoxazol-2-yl)-
r
-1,
t
-3-bis[4-(dimethylamino)phenyl]cyclobutane
Feng-Yu Li, Shu-Tao Wang, Jun-Peng Zhuang, Lei Jiang and Yan-Lin Song*
Key Laboratory of Organic Solids, Institute of Chemistry and Centre for Molecular Science, Chinese Academy of Sciences, Beijing 100080, People’s Republic of China
Correspondence e-mail: ylsong@iccas.ac.cn
Key indicators
Single-crystal X-ray study T= 293 K
Mean(C–C) = 0.005 A˚ Rfactor = 0.064 wRfactor = 0.218
Data-to-parameter ratio = 13.5
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
Received 29 December 2006 Accepted 30 January 2007
#2007 International Union of Crystallography All rights reserved
The title compound, C34H32N4O2, undergoes a reversible [2+2]
photoreaction with 1-(2-benzoxazol-2-yl)-2-bis[4-(dimethyl-amino)phenyl]ethane (NBE). Through intermolecular head-to-tail [2+2] photocycloaddition of NBE, the title compound displays a centrosymmetric molecular structure in the crystal-line state. The cyclobutane ring has an exactly planar conformation and is nearly square.
Comment
Utilizing the two distinct isomeric states of [2+2] photo-reaction between diarylethenes and tetraarylcyclobutanes, tetraarylcyclobutanes can be applied as optical data-storage materials. The molecular structure of tetraarylcyclobutane enables the study of the mechanism of action of such storage materials and the improvement of their properties. An average puckered conformation of the cyclobutane ring is found in tetraarylcyclobutanes such asr-1,c-2,t-3,t -4-1,3-bis[2-(4-R-phenyl)]-2,4-di(4-pyridyl)-cyclobutane, withR= Cl, CH3
and C6H5(Busettiet al., 1980; Zhang, Liet al., 2000). However,
a planar conformation of the cyclobutane ring has been observed in some tetraheteroaryl- and diphenylbisheteroaryl-substituted cyclobutanes, such as r-1,c-2,t-3,t -4-tetrakis-(2-R)cyclobutane, with R = 6,7-dimethylbenzoxazolyl, 5-phenyloxazolyl and benzoxazolyl (Kaoet al., 1989; Zhanget al., 1996; Zhang, Zhanget al., 2000), andr-1,c-2,t-3,t
-4-1,3-bis-R-2,4-bis(4-methoxyphenyl)cyclobutane, with R = 5-methyl-benzoxazolyl and 2-(5-phenyl-1,3,4-oxadiazolyl) (Zhanget al., 2001; Zhenget al., 2001). The title compound, (I), has a head-to-tail structure, and the cyclobutane ring has a planar conformation.
bond angles in the centrosymmetric cyclobutane ring of (I) are 90.8 (2) and 89.2 (2). The C—C bonds of the cyclobutane ring
are equal in length [1.577 (4) A˚ ], and although this is longer than a normal C—C single bond, this elongation is common for cyclobutanes (Zhenget al., 2001; Zhuanget al., 2002).
The benzoxazolyl group is planar, while the benzeneamine group is nearly planar, with a slight twist between the benzene and dimethylamine groups of 5.8 (7), demonstrating that the
lone electron pair of the N atom is not completely conjugated with thesystem of the aryl group when there is no acceptor group at the opposite position of the benzene ring. The dihedral angle between the benzoxazolyl and benzeneamine planes, which are situated on the same side of the cyclobutane ring, is 59.38 (12), and the dihedral angles between the
cyclobutane ring plane and the benzoxazolyl and benzene-amine planes are 87.50 (12) and 44.56 (11), respectively.
Experimental
A solution of 4-(dimethylamino)benzaldehyde hydrochloride (1.86 g, 0.01 mol) and 2-methylbenzooxazole (1.80 ml, 0.015 mol) in dimethylformamide (50 ml) was stirred at 353 K. KOH (5.0 g, 0.089 mol) was added to the reaction mixture over a period of 1 h. After 4 h, the mixture was added quickly to water (100 ml) and a precipitate was obtained. Recrystallization from ethanol gave yellow needle crystals of 1-(N,N -dimethyl-4-benzeneamino)-2-(2-benzoxazolyl)ethane (NBE) (2.2 g, 84%). A solution of NBE (50 mg) in acetonitrile (80 ml) was placed under a 300 W high-pressure
cooled in a refrigerator overnight and then filtered to isolate the precipitate. Recrystallization from a solution in a dimethyl-formamide-ethanol mixture (1:1) gave single crystals of (I).
Crystal data
C34H32N4O2
Mr= 528.64
Monoclinic,C2=c a= 24.441 (5) A˚
b= 6.0694 (12) A˚
c= 20.101 (4) A˚
= 108.46 (3)
V= 2828.4 (10) A˚3
Z= 4
Dx= 1.241 Mg m 3
MoKradiation
= 0.08 mm 1
T= 293 (2) K Rod, colourless 0.440.350.21 mm
Data collection
Rigaku R-AXIS RAPID image-plate diffractometer
!scans
Absorption correction: none 12682 measured reflections
2476 independent reflections 1549 reflections withI> 2(I)
Rint= 0.037
max= 25.0
Refinement
Refinement onF2
R[F2> 2(F2)] = 0.064
wR(F2) = 0.218
S= 1.02 2476 reflections 183 parameters
H-atom parameters constrained
w= 1/[2
(Fo2) + (0.1378P)2]
whereP= (Fo2+ 2Fc2)/3
(/)max< 0.001
max= 0.32 e A˚ 3
min= 0.41 e A˚ 3
Methyl H atoms were placed in calculated positions, with C—H = 0.96 A˚ , their torsion angles were refined to fit the electron density, andUiso(H) = 1.5Ueq(C). Other H atoms were placed in calculated positions, with C—H = 0.93 (aromatic) or 0.98 A˚ (methine), and refined in riding mode, withUiso(H) = 1.2Ueq(C).
Data collection:RAPID-AUTO (Rigaku, 1998); cell refinement:
RAPID-AUTO; data reduction: CrystalStructure (Rigaku/MSC, 2002); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure:SHELXL97(Sheldrick, 1997); molecular graphics:SHELXTL(Bruker, 2002); software used to prepare material for publication:SHELXTL.
We thank Dr Hong-Wei Ma of the Institute of Chemistry, Chinese Academy of Sciences, for collecting the intensity data.
References
Bruker (2002).SHELXTL. Bruker AXS Inc., Madison, Wisconsin, USA. Busetti, V., Valle, G., Zanotti, G. & Galiazzo, G. (1980).Acta Cryst.B36, 894–
897.
Kao, C. H., Zhou, Y. M., Xia, X. P., Wang, H. G. & Wang, R. J. (1989).J. Struct. Chem.8, 53–56.
Rigaku (1998).PROCESS-AUTO. Rigaku Corporation, Tokyo, Japan. Rigaku/MSC (2002).CrystalStructure. Version 3.00. Rigaku/MSC, The
Wood-lands, Texas, USA.
Sheldrick, G. M. (1997). SHELXS97 and SHELXL97. University of Go¨ttingen, Germany.
Zhang, W. Q., Li, S. L., Zhang, Z. M., Qi, X., Zheng, Y., Zhuang, J. P. & Mak, T. C. W. (2000).Chem. J. Chin. Univ.21, 556–561.
Zhang, W. Q., Wang, M. Z. & Wang, H. G. (1996).Sci. China Ser. B,39, 105– 112.
Zhang, W. Q., Zhang, Z. M., Zheng, Y., Wang, S. L. & Zhao, S. N. (2000).Acta Phys. Chim. Sin.16, 207–213.
Zhang, W. Q., Zhuang, J. P., Li, C. B., Sun, H. & Yuan, X. N. (2001).Chin. J. Chem. .19, 695–701.
[image:2.610.56.277.69.353.2]Zheng, Y., Zhuang, J.-P., Zhang, W.-Q., Leng, X.-B. & Weng, L.-H. (2001).Acta
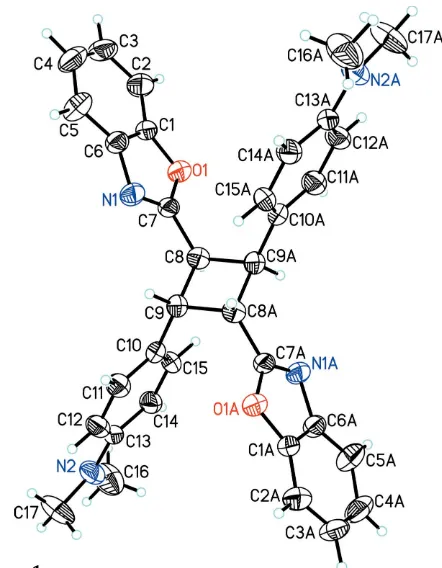
Figure 1
The molecular structure of (I), with 30% probability displacement ellipsoids (arbitrary spheres for the H atoms). [Symmetry code: (A)1
2 x, 3
supporting information
sup-1 Acta Cryst. (2007). E63, o1171–o1172
supporting information
Acta Cryst. (2007). E63, o1171–o1172 [https://doi.org/10.1107/S1600536807004813]
c
-2,
t
-4-Bis(2-benzoxazol-2-yl)-
r
-1,
t
-3-bis[4-(dimethylamino)phenyl]cyclobutane
Feng-Yu Li, Shu-Tao Wang, Jun-Peng Zhuang, Lei Jiang and Yan-Lin Song
r-1,c-2,t-3,t-4–1,3-bis(N,N-dimethyl-4-benzenamino)-2,4-bis- (2-benzooxzolyl)cyclobutane
Crystal data
C34H32N4O2 Mr = 528.64
Monoclinic, C2/c
Hall symbol: -C 2yc
a = 24.441 (5) Å
b = 6.0694 (12) Å
c = 20.101 (4) Å
β = 108.46 (3)°
V = 2828.4 (10) Å3
Z = 4
F(000) = 1120
Dx = 1.241 Mg m−3
Melting point: 548(1) K
Mo Kα radiation, λ = 0.71073 Å
Cell parameters from 12682 reflections
θ = 2.3–25.0°
µ = 0.08 mm−1
T = 293 K
Platelet, colourless 0.44 × 0.35 × 0.21 mm
Data collection
Rigaku R-AXIS RAPID image-plate diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
ω scans
12682 measured reflections 2476 independent reflections
1549 reflections with I > 2σ(I)
Rint = 0.037
θmax = 25.0°, θmin = 1.7°
h = −28→28
k = −7→7
l = −23→22
Refinement
Refinement on F2
Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.064 wR(F2) = 0.218
S = 1.02
2476 reflections 183 parameters 18 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained
w = 1/[σ2(F
o2) + (0.1378P)2]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.32 e Å−3
Δρmin = −0.41 e Å−3
Special details
Experimental. 1-(N,N-dimethyl-4-benzenamino)-2-(2-benzooxazolyl)ethane: 1H NMR (300 MHz, CDCl
3, δ, p.p.m.):
7.76–7.26 (7H, m), 6.88–6.71 (3H, m), 3.03 (6H, s); 13C NMR (300 MHz, CDCl
3, δ, p.p.m.): 163.2, 150.6, 149.6, 141.7,
139.2, 128.3, 124.1, 123.7, 118.6, 111.3, 109.3, 107.8, 39.5.
The title compound: 1H NMR (300 MHz, CDCl
3, δ, p.p.m.): 7.65–7.18 (6H, m), 6.53 (2H, d), 4.93 (1H, t), 4.77 (1H, t),
2.80 (6H, s); 13C NMR (300 MHz, CDCl
3, δ, p.p.m.): 165.1, 150.2, 148.7, 140.4, 127.3, 123.5, 123.0, 118.8, 111.8, 109.5,
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
N1 0.12010 (11) 0.8052 (4) 0.42964 (14) 0.0633 (7)
O1 0.14612 (9) 1.1341 (4) 0.47938 (11) 0.0674 (6)
N2 0.36330 (13) 0.8422 (5) 0.24634 (14) 0.0779 (9)
C1 0.08670 (12) 1.1275 (5) 0.45478 (15) 0.0571 (8)
C2 0.04860 (16) 1.2903 (7) 0.4566 (2) 0.0839 (11)
H2A 0.0606 1.4271 0.4768 0.101*
C3 −0.00897 (17) 1.2375 (8) 0.4265 (2) 0.0902 (12)
H3A −0.0367 1.3419 0.4268 0.108*
C4 −0.02618 (16) 1.0406 (8) 0.3967 (2) 0.0902 (12)
H4A −0.0655 1.0124 0.3775 0.108*
C5 0.01267 (15) 0.8786 (7) 0.3937 (2) 0.0877 (11)
H5A 0.0006 0.7439 0.3720 0.105*
C6 0.07010 (13) 0.9273 (5) 0.42462 (15) 0.0598 (8)
C7 0.16147 (12) 0.9337 (5) 0.46173 (14) 0.0547 (7)
C8 0.22512 (12) 0.8992 (5) 0.48027 (14) 0.0580 (8)
H8A 0.2444 1.0338 0.4724 0.070*
C9 0.24309 (12) 0.7011 (5) 0.44503 (14) 0.0560 (8)
H9A 0.2090 0.6119 0.4217 0.067*
C10 0.27671 (11) 0.7423 (5) 0.39464 (14) 0.0537 (7)
C11 0.27387 (14) 0.5949 (6) 0.34313 (17) 0.0664 (9)
H11A 0.2516 0.4687 0.3399 0.080*
C12 0.30281 (14) 0.6255 (5) 0.29555 (17) 0.0675 (9)
H12A 0.3000 0.5182 0.2616 0.081*
C13 0.33623 (12) 0.8109 (5) 0.29618 (13) 0.0528 (7)
C14 0.34013 (13) 0.9618 (5) 0.34893 (16) 0.0645 (8)
H14A 0.3627 1.0873 0.3523 0.077*
C15 0.31056 (13) 0.9282 (6) 0.39711 (15) 0.0666 (9)
H15A 0.3136 1.0330 0.4318 0.080*
C16 0.3976 (2) 1.0327 (8) 0.2478 (2) 0.1156 (17)
H16A 0.3761 1.1620 0.2517 0.173*
H16B 0.4076 1.0401 0.2054 0.173*
H16C 0.4321 1.0247 0.2874 0.173*
C17 0.3640 (2) 0.6713 (8) 0.1978 (2) 0.1190 (17)
H17A 0.3259 0.6117 0.1783 0.179*
H17B 0.3899 0.5568 0.2217 0.179*
supporting information
sup-3 Acta Cryst. (2007). E63, o1171–o1172
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
N1 0.0584 (16) 0.0588 (15) 0.0689 (15) 0.0188 (13) 0.0146 (13) 0.0009 (13)
O1 0.0541 (13) 0.0771 (14) 0.0686 (13) 0.0112 (11) 0.0159 (10) −0.0074 (11)
N2 0.0839 (19) 0.097 (2) 0.0650 (16) −0.0009 (17) 0.0405 (15) −0.0067 (15)
C1 0.0479 (17) 0.074 (2) 0.0506 (16) 0.0117 (15) 0.0171 (13) 0.0014 (15)
C2 0.075 (2) 0.088 (3) 0.088 (2) 0.025 (2) 0.024 (2) −0.012 (2)
C3 0.065 (2) 0.114 (3) 0.095 (3) 0.041 (2) 0.031 (2) 0.014 (3)
C4 0.0455 (19) 0.117 (3) 0.102 (3) 0.012 (2) 0.0138 (19) 0.022 (3)
C5 0.052 (2) 0.090 (3) 0.107 (3) 0.0029 (19) 0.0065 (19) 0.004 (2)
C6 0.0522 (18) 0.070 (2) 0.0568 (17) 0.0109 (16) 0.0171 (14) 0.0099 (16)
C7 0.0494 (17) 0.0718 (19) 0.0456 (15) 0.0140 (16) 0.0187 (13) 0.0099 (15)
C8 0.0575 (18) 0.0590 (16) 0.0585 (17) 0.0032 (14) 0.0200 (14) −0.0032 (14)
C9 0.0506 (16) 0.0630 (18) 0.0513 (15) −0.0011 (14) 0.0116 (13) −0.0087 (14)
C10 0.0409 (14) 0.0688 (18) 0.0465 (14) 0.0040 (14) 0.0070 (11) −0.0022 (14)
C11 0.067 (2) 0.0685 (19) 0.0654 (19) −0.0003 (16) 0.0230 (16) −0.0077 (17)
C12 0.070 (2) 0.073 (2) 0.0643 (18) 0.0007 (17) 0.0280 (16) −0.0183 (16)
C13 0.0476 (15) 0.0653 (18) 0.0449 (15) 0.0118 (14) 0.0141 (12) −0.0010 (14)
C14 0.0579 (19) 0.072 (2) 0.0640 (18) −0.0064 (16) 0.0204 (15) −0.0140 (17)
C15 0.0614 (19) 0.086 (2) 0.0489 (15) 0.0082 (17) 0.0133 (13) −0.0220 (16)
C16 0.140 (4) 0.129 (4) 0.100 (3) −0.037 (3) 0.070 (3) −0.002 (3)
C17 0.165 (5) 0.124 (4) 0.101 (3) −0.007 (3) 0.089 (3) −0.033 (3)
Geometric parameters (Å, º)
N1—C7 1.278 (4) C8—H8A 0.9800
N1—C6 1.405 (4) C9—C10 1.514 (4)
O1—C7 1.353 (4) C9—C8i 1.578 (4)
O1—C1 1.379 (4) C9—H9A 0.9800
N2—C13 1.378 (4) C10—C11 1.354 (4)
N2—C16 1.423 (5) C10—C15 1.390 (4)
N2—C17 1.428 (5) C11—C12 1.370 (4)
C1—C6 1.361 (4) C11—H11A 0.9300
C1—C2 1.366 (4) C12—C13 1.388 (4)
C2—C3 1.382 (6) C12—H12A 0.9300
C2—H2A 0.9300 C13—C14 1.382 (4)
C3—C4 1.343 (6) C14—C15 1.395 (4)
C3—H3A 0.9300 C14—H14A 0.9300
C4—C5 1.381 (5) C15—H15A 0.9300
C4—H4A 0.9300 C16—H16A 0.9600
C5—C6 1.375 (5) C16—H16B 0.9600
C5—H5A 0.9300 C16—H16C 0.9600
C7—C8 1.495 (4) C17—H17A 0.9600
C8—C9 1.529 (4) C17—H17B 0.9600
C8—C9i 1.578 (4) C17—H17C 0.9600
C7—O1—C1 103.2 (2) C10—C9—H9A 109.3
C13—N2—C16 120.8 (3) C8—C9—H9A 109.3
C13—N2—C17 120.6 (3) C8i—C9—H9A 109.3
C16—N2—C17 118.1 (3) C11—C10—C15 116.7 (3)
C6—C1—C2 123.2 (3) C11—C10—C9 119.7 (3)
C6—C1—O1 108.5 (2) C15—C10—C9 123.6 (3)
C2—C1—O1 128.2 (3) C10—C11—C12 122.1 (3)
C1—C2—C3 115.4 (4) C10—C11—H11A 119.0
C1—C2—H2A 122.3 C12—C11—H11A 119.0
C3—C2—H2A 122.3 C11—C12—C13 122.4 (3)
C4—C3—C2 122.2 (3) C11—C12—H12A 118.8
C4—C3—H3A 118.9 C13—C12—H12A 118.8
C2—C3—H3A 118.9 N2—C13—C14 122.3 (3)
C3—C4—C5 122.0 (4) N2—C13—C12 121.4 (3)
C3—C4—H4A 119.0 C14—C13—C12 116.2 (3)
C5—C4—H4A 119.0 C13—C14—C15 120.7 (3)
C6—C5—C4 116.4 (4) C13—C14—H14A 119.6
C6—C5—H5A 121.8 C15—C14—H14A 119.6
C4—C5—H5A 121.8 C10—C15—C14 121.8 (3)
C1—C6—C5 120.7 (3) C10—C15—H15A 119.1
C1—C6—N1 108.0 (3) C14—C15—H15A 119.1
C5—C6—N1 131.3 (3) N2—C16—H16A 109.5
N1—C7—O1 116.1 (2) N2—C16—H16B 109.5
N1—C7—C8 129.4 (3) H16A—C16—H16B 109.5
O1—C7—C8 114.4 (3) N2—C16—H16C 109.5
C7—C8—C9 115.0 (3) H16A—C16—H16C 109.5
C7—C8—C9i 116.8 (2) H16B—C16—H16C 109.5
C9—C8—C9i 90.6 (2) N2—C17—H17A 109.5
C7—C8—H8A 111.0 N2—C17—H17B 109.5
C9—C8—H8A 111.0 H17A—C17—H17B 109.5
C9i—C8—H8A 111.0 N2—C17—H17C 109.5
C10—C9—C8 118.4 (3) H17A—C17—H17C 109.5
C10—C9—C8i 119.5 (2) H17B—C17—H17C 109.5
C7—O1—C1—C6 0.2 (3) C7—C8—C9—C10 116.2 (3)
C7—O1—C1—C2 −177.8 (3) C9i—C8—C9—C10 −123.7 (3)
C6—C1—C2—C3 1.0 (5) C7—C8—C9—C8i −120.2 (3)
O1—C1—C2—C3 178.8 (3) C9i—C8—C9—C8i 0.0
C1—C2—C3—C4 −0.7 (6) C8—C9—C10—C11 −153.9 (3)
C2—C3—C4—C5 −0.7 (6) C8i—C9—C10—C11 99.1 (3)
C3—C4—C5—C6 1.7 (6) C8—C9—C10—C15 25.1 (4)
C2—C1—C6—C5 0.1 (5) C8i—C9—C10—C15 −81.9 (4)
O1—C1—C6—C5 −178.1 (3) C15—C10—C11—C12 0.1 (5)
C2—C1—C6—N1 177.6 (3) C9—C10—C11—C12 179.1 (3)
O1—C1—C6—N1 −0.6 (3) C10—C11—C12—C13 −1.0 (5)
C4—C5—C6—C1 −1.4 (5) C16—N2—C13—C14 0.9 (5)
C4—C5—C6—N1 −178.3 (3) C17—N2—C13—C14 172.5 (4)
supporting information
sup-5 Acta Cryst. (2007). E63, o1171–o1172
C7—N1—C6—C5 177.8 (3) C17—N2—C13—C12 −8.2 (5)
C6—N1—C7—O1 −0.6 (3) C11—C12—C13—N2 −177.6 (3)
C6—N1—C7—C8 −177.6 (3) C11—C12—C13—C14 1.7 (5)
C1—O1—C7—N1 0.2 (3) N2—C13—C14—C15 177.8 (3)
C1—O1—C7—C8 177.7 (2) C12—C13—C14—C15 −1.5 (4)
N1—C7—C8—C9 11.8 (4) C11—C10—C15—C14 0.1 (5)
O1—C7—C8—C9 −165.3 (2) C9—C10—C15—C14 −178.9 (3)
N1—C7—C8—C9i −92.6 (3) C13—C14—C15—C10 0.7 (5)
O1—C7—C8—C9i 90.3 (3)