Genetic and Physical Mapping of Genic Microsatellites in Barley (Hordeum vulgare L )
Full text
Figure


Related documents
The objectives of our study were to (i) examine the population structure and the genetic diversity in elite maize germplasm based on simple sequence repeat (SSR)
We have tested the efficacy of putative microsatellite single sequence repeat SSR markers, previously identified in a 2-49 Gluyas Early/Gala × Janz doubled haploid wheat Triticum
A genetic map of barley with 224 AFLP and 39 simple sequence repeat SSR markers was constructed using a doubled haploid DH mapping population from a cross between the varieties
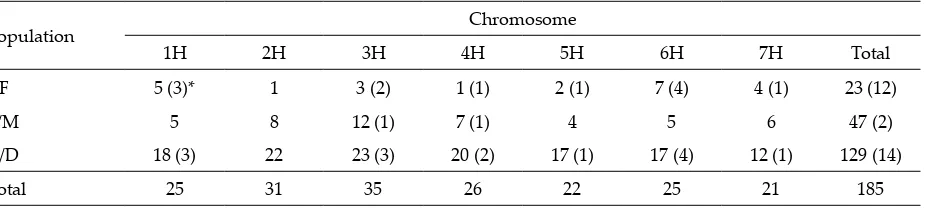
Mapping of aligned markers to barley genetic/physical maps 216 . Markers aligned to sequence DBs (Table 1) were then assigned
TAM (random wheat genomic and cDNA clones) CSIH (random T.. Markers preceded by an X were mapped with DNA probes. The function of the markers not preceded by an X
Simple sequence repeat markers (SSR) are being extensively used in genome studies, marker-assisted selection, and cultivar identification. These are also well-known for
When the varieties were grouped using markers with above-average Polymorphic Information Content (PIC) values, the same groups were obtained as when using all markers, outlining
(1987) found that grain yield in barley was significantly correlated with plant height and spike length and that these two components had high positive