organic papers
Acta Cryst.(2006). E62, o871–o872 doi:10.1107/S1600536806001735 Wanget al. C
11H12O3
o871
Acta Crystallographica Section E Structure Reports Online
ISSN 1600-5368
(
E
)-4-(4-Hydroxy-3-methoxyphenyl)but-3-en-2-one
Shi-Fan Wang,a* Li-Jun Zhoub and Xian Yangb
a
Department of Pharmaceutical Engineering, School of Ocean, Hainan University, Haikou 570228, People’s Republic of China, and
bLaboratory for Drug Design and Synthesis,
School of Ocean, Hainan University, Haikou 570228, People’s Republic of China
Correspondence e-mail: wangsf777@163.com
Key indicators
Single-crystal X-ray study
T= 293 K
Mean(C–C) = 0.002 A˚
Rfactor = 0.046
wRfactor = 0.140
Data-to-parameter ratio = 18.0
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
Received 30 November 2005 Accepted 16 January 2006
#2006 International Union of Crystallography
All rights reserved
The title compound, C11H12O3, was synthesized from
4-hydroxy-3-methoxybenzaldehyde and acetone. The molecule has a high degree of conjugation throughout the system and intermolecular hydrogen bonds connect adjacent molecules to form one-dimensional chains.
Comment
,-Unsaturated ketones are an important class of pharma-ceutical intermediates and have extensive application in the synthesis of natural products and drugs by 1,4 addition. As a precursor for further synthesis, we have synthesized the title compound, (I).
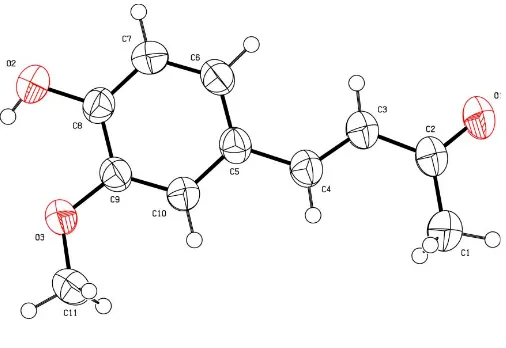
All the bond lengths and angles (Table 1) are within normal ranges (Allenet al., 1987) and the structural data confirm the
E configuration about the C3 C4 double bond. Atoms C2, C3, C4 and O1 constitute a well defined plane and the benzene ring plane is inclined at 5.34 (1)to this plane (Fig. 1). A weak
intermolecular O2—H2 O1 hydrogen bond links adjacent molecules, forming one-dimensional chains (Fig. 2).
Experimental
4-Hydroxy-3-methoxybenzaldehyde (3.04 g, 20 mmol) was dissolved in 25 ml of acetone and 12 ml of dilute aqueous NaOH solution (10%) was added to the acetone solution with stirring. The mixture was allowed to stand overnight at room temperature and then the mixture was acidified with dilute aqueous HCl to give (E )-4-(4-hydroxy-3-methoxyphenyl)but-3-en-2-one as a yellow solid (yield 73%). The resultant precipitate was filtered off, washed with water and recrystallized from ethanol and dichloromethane (2:1), in air over a period of four days. After about three-quarters of the original solvent had evaporated, large colourless prisms of (I) were obtained (yield 61%).
Crystal data
C11H12O3 Mr= 192.21
Monoclinic,P21=c a= 9.602 (5) A˚ b= 7.780 (4) A˚ c= 13.478 (7) A˚ = 97.466 (8)
V= 998.2 (9) A˚3 Z= 4
Dx= 1.279 Mg m
3 MoKradiation Cell parameters from 2995
reflections = 2.6–27.9
Data collection
Bruker SMART CCD area-detector diffractometer
’and!scans
Absorption correction: none 8188 measured reflections 2355 independent reflections
1816 reflections withI> 2(I) Rint= 0.020
max= 28.4 h=12!12 k=10!10 l=17!17
Refinement
Refinement onF2 R[F2> 2(F2)] = 0.046 wR(F2) = 0.140 S= 1.04 2355 reflections 131 parameters
H-atom parameters constrained
w= 1/[2
(Fo2) + (0.07P)2 + 0.1629P]
whereP= (Fo2+ 2Fc2)/3 (/)max= 0.001
max= 0.20 e A˚
3
min=0.19 e A˚
3
[image:2.610.315.568.70.244.2]Extinction correction:SHELXL97 Extinction coefficient: 0.024 (5)
Table 1
Selected geometric parameters (A˚ ,).
O2—C8 1.3556 (18)
C4—C3 1.336 (2)
C4—C5 1.459 (2)
C5—C6 1.388 (2)
C5—C10 1.400 (2)
O3—C9 1.3686 (17)
O3—C11 1.413 (2)
C10—C9 1.3785 (19)
C8—C7 1.380 (2)
C8—C9 1.394 (2)
C6—C7 1.379 (2)
O1—C2 1.2243 (18)
C3—C2 1.454 (2)
C2—C1 1.490 (2)
C3—C4—C5 127.19 (15)
C6—C5—C10 118.55 (12)
C6—C5—C4 122.60 (13)
C10—C5—C4 118.85 (13)
C9—O3—C11 117.63 (12)
C9—C10—C5 120.79 (13)
O2—C8—C7 118.81 (13)
O2—C8—C9 121.70 (13)
C7—C8—C9 119.48 (13)
O3—C9—C10 125.48 (13)
O3—C9—C8 114.63 (12)
C10—C9—C8 119.89 (13)
C7—C6—C5 120.63 (14)
C4—C3—C2 124.57 (15)
O1—C2—C3 119.85 (15)
O1—C2—C1 119.74 (14)
C3—C2—C1 120.40 (14)
C6—C7—C8 120.64 (14)
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
O2—H2 O1i 0.82 2.08 2.772 (2) 141
Symmetry code: (i)xþ1;yþ1 2;z
1 2.
H atoms were positioned geometrically [C—H distances of 0.93 (Csp2—H) and 0.96 A˚ (methyl), and O—H = 0.82 A˚].Uiso(H) values
were set equal toxUeq(carrier atom), wherex= 1.2 for CH and 1.5 for
O and methyl C.
Data collection:SMART(Bruker, 2002); cell refinement:SAINT
(Bruker, 2002); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics:
SHELXTL(Sheldrick, 2000); software used to prepare material for publication:SHELXTL.
This project is sponsored by the Scientific Research Foundation of Hainan University, China.
References
Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987).J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
Bruker (2002).SAINTandSMART. Bruker AXS Inc., Madison, Wisconsin, USA.
Sheldrick, G. M. (1997). SHELXS97 and SHELXL97. University of Go¨ttingen, Germany.
Sheldrick, G. M. (2000).SHELXTL. Version 6.10. Bruker AXS Inc., Madison, Wisconsin, USA.
Figure 1
[image:2.610.314.563.286.359.2]The structure of the title compound (1), showing 50% probability displacement ellipsoids and the atom-numbering scheme.
Figure 2
[image:2.610.44.299.300.450.2]supporting information
sup-1 Acta Cryst. (2006). E62, o871–o872
supporting information
Acta Cryst. (2006). E62, o871–o872 [https://doi.org/10.1107/S1600536806001735]
(
E
)-4-(4-Hydroxy-3-methoxyphenyl)but-3-en-2-one
Shi-Fan Wang, Li-Jun Zhou and Xian Yang
(E)-4-(4-hydroxy-3-methoxyphenyl) but-3-en-2-one
Crystal data
C11H12O3
Mr = 192.21
Monoclinic, P21/c
a = 9.602 (5) Å
b = 7.780 (4) Å
c = 13.478 (7) Å
β = 97.466 (8)°
V = 998.2 (9) Å3
Z = 4
F(000) = 408
Dx = 1.279 Mg m−3
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 2995 reflections
θ = 2.6–27.9°
µ = 0.09 mm−1
T = 293 K Prism, colourless 0.36 × 0.28 × 0.22 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
φ and ω scans
8188 measured reflections 2355 independent reflections
1816 reflections with I > 2σ(I)
Rint = 0.020
θmax = 28.4°, θmin = 2.1°
h = −12→12
k = −10→10
l = −17→17
Refinement
Refinement on F2
Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.046
wR(F2) = 0.140
S = 1.04 2355 reflections 131 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained
w = 1/[σ2(F
o2) + (0.07P)2 + 0.1629P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max = 0.001
Δρmax = 0.20 e Å−3
Δρmin = −0.19 e Å−3
Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4
Extinction coefficient: 0.024 (5)
Special details
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
O2 1.20118 (12) 0.80293 (18) 0.28139 (8) 0.0676 (4)
H2 1.2674 0.7414 0.2716 0.101*
C4 0.80091 (15) 0.82090 (19) −0.06027 (11) 0.0519 (4)
H4 0.8317 0.7829 −0.1191 0.062*
C5 0.90466 (13) 0.82078 (18) 0.02899 (10) 0.0471 (3) O3 1.26019 (11) 0.64976 (17) 0.11386 (8) 0.0683 (4) C10 1.03328 (14) 0.73781 (19) 0.02537 (10) 0.0494 (4)
H10 1.0519 0.6873 −0.0340 0.059*
C8 1.10596 (14) 0.80697 (19) 0.19783 (10) 0.0491 (4) C9 1.13266 (14) 0.73009 (19) 0.10883 (10) 0.0481 (3) C6 0.88071 (15) 0.8990 (2) 0.11778 (11) 0.0536 (4)
H6 0.7965 0.9563 0.1212 0.064*
O1 0.44087 (11) 0.88424 (18) −0.15226 (9) 0.0725 (4) C3 0.66655 (15) 0.8696 (2) −0.06681 (12) 0.0539 (4)
H3 0.6358 0.9171 −0.0102 0.065*
C2 0.56503 (15) 0.8534 (2) −0.15629 (12) 0.0550 (4) C7 0.98065 (16) 0.8924 (2) 0.20102 (11) 0.0564 (4)
H7 0.9635 0.9462 0.2599 0.068*
C11 1.31211 (18) 0.6130 (3) 0.02279 (13) 0.0686 (5)
H11A 1.3154 0.7170 −0.0152 0.103*
H11B 1.2513 0.5316 −0.0148 0.103*
H11C 1.4049 0.5656 0.0366 0.103*
C1 0.61088 (19) 0.7969 (3) −0.25261 (14) 0.0864 (7)
H1A 0.5309 0.7915 −0.3033 0.130*
H1B 0.6534 0.6853 −0.2442 0.130*
H1C 0.6778 0.8775 −0.2724 0.130*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
supporting information
sup-3 Acta Cryst. (2006). E62, o871–o872
C2 0.0413 (8) 0.0594 (9) 0.0625 (9) −0.0014 (6) −0.0004 (6) 0.0014 (7) C7 0.0564 (8) 0.0674 (10) 0.0464 (8) 0.0073 (7) 0.0098 (6) −0.0028 (7) C11 0.0530 (9) 0.0917 (13) 0.0630 (10) 0.0170 (8) 0.0150 (7) 0.0022 (9) C1 0.0538 (10) 0.1337 (19) 0.0680 (11) 0.0088 (11) −0.0059 (8) −0.0250 (12)
Geometric parameters (Å, º)
O2—C8 1.3556 (18) C6—C7 1.379 (2)
O2—H2 0.8200 C6—H6 0.9300
C4—C3 1.336 (2) O1—C2 1.2243 (18)
C4—C5 1.459 (2) C3—C2 1.454 (2)
C4—H4 0.9300 C3—H3 0.9300
C5—C6 1.388 (2) C2—C1 1.490 (2)
C5—C10 1.400 (2) C7—H7 0.9300
O3—C9 1.3686 (17) C11—H11A 0.9600
O3—C11 1.413 (2) C11—H11B 0.9600
C10—C9 1.3785 (19) C11—H11C 0.9600
C10—H10 0.9300 C1—H1A 0.9600
C8—C7 1.380 (2) C1—H1B 0.9600
C8—C9 1.394 (2) C1—H1C 0.9600
C8—O2—H2 109.5 C4—C3—H3 117.7
C3—C4—C5 127.19 (15) C2—C3—H3 117.7
C3—C4—H4 116.4 O1—C2—C3 119.85 (15)
C5—C4—H4 116.4 O1—C2—C1 119.74 (14)
C6—C5—C10 118.55 (12) C3—C2—C1 120.40 (14) C6—C5—C4 122.60 (13) C6—C7—C8 120.64 (14)
C10—C5—C4 118.85 (13) C6—C7—H7 119.7
C9—O3—C11 117.63 (12) C8—C7—H7 119.7
C9—C10—C5 120.79 (13) O3—C11—H11A 109.5
C9—C10—H10 119.6 O3—C11—H11B 109.5
C5—C10—H10 119.6 H11A—C11—H11B 109.5
O2—C8—C7 118.81 (13) O3—C11—H11C 109.5 O2—C8—C9 121.70 (13) H11A—C11—H11C 109.5 C7—C8—C9 119.48 (13) H11B—C11—H11C 109.5 O3—C9—C10 125.48 (13) C2—C1—H1A 109.5
O3—C9—C8 114.63 (12) C2—C1—H1B 109.5
C10—C9—C8 119.89 (13) H1A—C1—H1B 109.5
C7—C6—C5 120.63 (14) C2—C1—H1C 109.5
C7—C6—H6 119.7 H1A—C1—H1C 109.5
C5—C6—H6 119.7 H1B—C1—H1C 109.5
C4—C3—C2 124.57 (15)
C11—O3—C9—C8 −162.13 (15) C4—C3—C2—O1 172.18 (17) C5—C10—C9—O3 178.73 (14) C4—C3—C2—C1 −6.9 (3) C5—C10—C9—C8 −0.6 (2) C5—C6—C7—C8 −0.5 (2) O2—C8—C9—O3 0.9 (2) O2—C8—C7—C6 −179.72 (14) C7—C8—C9—O3 179.65 (13) C9—C8—C7—C6 1.5 (2)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
O2—H2···O1i 0.82 2.08 2.772 (2) 141