metal-organic papers
Acta Cryst.(2006). E62, m45–m46 doi:10.1107/S1600536805040249 Zhong-Lu You [Cu(C
13H18BrN2O)(NCS)]
m45
Acta Crystallographica Section E Structure Reports
Online
ISSN 1600-5368
{4-Bromo-2-[(2-diethylaminoethylimino)methyl]-phenolato}thiocyanatocopper(II)
Zhong-Lu You
Department of Chemistry and Chemical Engineering, Liaoning Normal University, Dalian 116029, People’s Republic of China
Correspondence e-mail: youzhonglu@yahoo.com.cn
Key indicators
Single-crystal X-ray study
T= 298 K
Mean(C–C) = 0.004 A˚
Rfactor = 0.036
wRfactor = 0.091
Data-to-parameter ratio = 18.6
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
#2006 International Union of Crystallography Printed in Great Britain – all rights reserved
The title compound, [Cu(C13H18BrN2O)(NCS)], is a
mono-nuclear Schiff base copper(II) complex. The CuII atom is coordinated by one O and two N atoms of the Schiff base ligand, and by one N atom of the thiocyanate ligand, forming a square-planar coordination.
Comment
Recently, we have reported a series of Schiff base complexes (You, 2005a,b,c). As an extension of this work on the struc-tural characterization of Schiff base complexes, we describe here the synthesis and structure of the title new copper(II) compound, (I).
[image:1.610.271.393.321.393.2]The molecular structure of compound (I) is illustrated in Fig. 1, and selected bond distances and angles are given in Table 1. Compound (I) is structurally similar to the copper(II) compounds reported recently (You, 2005d,e). The CuIIatom is four-coordinated, in a square-planar arrangement, by one O and two N atoms of the Schiff base ligand, and by one N atom of the thiocyanate anion. The values of thetransangles in the CuON3 square plane are 176.48 (12) and 176.09 (10),
indi-cating a slightly distorted square-planar coordination. The Cu—O and Cu—N bond lengths (Table 1) are comparable with the corresponding values observed in other Schiff base copper(II) complexes (You & Zhu, 2004) and, as expected, the
[image:1.610.208.461.585.723.2]Received 30 November 2005 Accepted 2 December 2005 Online 7 December 2005
Figure 1
bond involving amine atom N2 is longer than that involving imine atom N1 (Mondalet al., 2001).
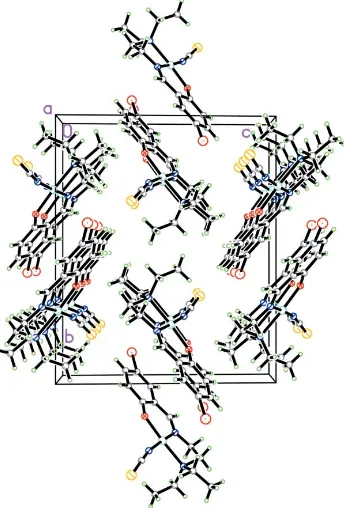
In the crystal structure, the molecules stack along theaaxis; the crystal packing is shown in Fig. 2.
Experimental
5-Bromosalicylaldehyde (0.1 mmol, 20.1 mg) and N,N0 -diethyl-ethane-1,2-diamine (0.1 mmol, 11.6 mg) were dissolved in MeOH (10 ml). The mixture was stirred at room temperature for 20 min to give a yellow solution. To this solution were added an aqueous solution (2 ml) of NH4NCS (0.1 mmol, 6.5 mg) and a MeOH solution (3 ml) of Cu(CH3COO)2H2O (0.1 mmol, 19.9 mg), with stirring. The mixture was stirred for a further 20 min at room temperature and then filtered. The filtrate was kept in air for 5 d, during which time blue block-shaped crystals of (I) were formed.
Crystal data
[Cu(C13H18BrN2O)(NCS)]
Mr= 419.82 Monoclinic,P21=c a= 7.052 (1) A˚
b= 16.688 (2) A˚
c= 13.775 (2) A˚
= 94.79 (1) V= 1615.4 (4) A˚3
Z= 4
Dx= 1.726 Mg m
3 MoKradiation Cell parameters from 3328
reflections
= 2.4–24.6
= 3.96 mm1
T= 298 (2) K Block, blue
0.250.180.17 mm
Bruker SMART CCD area-detector diffractometer
!scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin= 0.404,Tmax= 0.510 11427 measured reflections
3564 independent reflections 2787 reflections withI> 2(I)
Rint= 0.031
max= 27.5
h=8!9
k=21!21
l=16!17
Refinement
Refinement onF2
R[F2> 2(F2)] = 0.036
wR(F2) = 0.091
S= 1.03 3564 reflections 192 parameters
H-atom parameters constrained
w= 1/[2(F
o2) + (0.0453P)2 + 0.4325P]
whereP= (Fo2+ 2Fc2)/3 (/)max< 0.001
max= 0.54 e A˚
3
min=0.41 e A˚
3
Table 1
Selected geometric parameters (A˚ ,).
Cu1—O1 1.902 (2)
Cu1—N1 1.928 (3)
Cu1—N3 1.938 (3)
Cu1—N2 2.103 (2)
O1—Cu1—N1 92.93 (9)
O1—Cu1—N3 89.52 (11)
N1—Cu1—N3 176.48 (12)
O1—Cu1—N2 176.09 (10)
N1—Cu1—N2 84.27 (10)
N3—Cu1—N2 93.41 (11)
The H atoms were placed in idealized positions and constrained to ride on their parent atoms, with C—H distances in the range 0.93– 0.97 A˚ , and withUiso(H) = 1.2 or 1.5Ueq(C).
Data collection:SMART(Bruker, 1998); cell refinement:SAINT (Bruker, 1998); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 1997a); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997a); molecular graphics: SHELXTL(Sheldrick, 1997b); software used to prepare material for publication:SHELXTL.
The author thanks Liaoning Normal University, People’s Republic of China, for funding this study.
References
Bruker (1998).SMART(Version 5.628) andSAINT(Version 6.02). Bruker AXS Inc., Madison, Wisconsin, USA.
Mondal, N., Mitra, S., Gramilich, V., Ghodsi, S. O. & Malik, K. M. A. (2001).
Polyhedron,20, 135–141.
Sheldrick, G. M. (1996).SADABS. University of Go¨ttingen, Germany. Sheldrick, G. M. (1997a). SHELXL97 and SHELXS97. University of
Go¨ttingen, Germany.
Sheldrick, G. M. (1997b). SHELXTL. (Version 5.10). Bruker AXS Inc., Madison, Wisconsin, USA.
You, Z.-L. (2005a).Acta Cryst.E61, m1571–m1573. You, Z.-L. (2005b).Acta Cryst.E61, m1601–m1603. You, Z.-L. (2005c).Acta Cryst.E61, m1637–m1638. You, Z.-L. (2005d).Acta Cryst.E61, m2226–m2227. You, Z.-L. (2005e).Acta Cryst.E61, m1963–m1964.
[image:2.610.87.259.74.328.2]You, Z.-L. & Zhu, H.-L. (2004).Z. Anorg. Allg. Chem.630, 2754–2760. Figure 2
supporting information
sup-1
Acta Cryst. (2006). E62, m45–m46supporting information
Acta Cryst. (2006). E62, m45–m46 [doi:10.1107/S1600536805040249]
{4-Bromo-2-[(2-diethylaminoethylimino)methyl]phenolato}thiocyanato-copper(II)
Zhong-Lu You
S1. Comment
Recently, we have reported a series of Schiff base complexes (You, 2005a,b,c). As an extension of this work on the
structural characterization of Schiff base complexes, we describe here the synthesis and structure of the title new
copper(II) compound, (I).
The molecular structure of compound (I) is illustrated in Fig. 1, and selected bond distances and angles are given in
Table 1. Compound (I) is structurally similar to the copper(II) compounds reported recently (You, 2005d,e). The CuII
atom is four-coordinated, in a square-planar arrangement, by one O and two N atoms of the Schiff base ligand, and by
one N atom of the thiocyanate anion. The values of the trans angles in the CuON3 square plane are 176.48 (12) and
176.09 (10)°, indicating a slightly distorted square-planar coordination. The Cu—O and Cu—N bond lengths (Table 1)
are comparable with the corresponding values observed in other Schiff base copper(II) complexes (You & Zhu, 2004)
and, as expected, the bond involving amine atom N2 [2.071 (3) Å] is longer than that involving imine atom N1 [1.925 (3)
Å] (Mondal et al., 2001).
In the crystal strucrure, the molecules stack along the a axis; the crystal packing is shown in Fig. 2.
S2. Experimental
5-Bromosalicylaldehy (0.1 mmol, 20.1 mg) and N,N′-diethylethane-1,2-diamine (0.1 mmol, 11.6 mg) were dissolved in
MeOH (10 ml). The mixture was stirred at room temperature for 20 min to give a yellow solution. To this solution were
added an aqueous solution (2 ml) of NH4NCS (0.1 mmol, 6.5 mg) and a MeOH solution (3 ml) of Cu(CH3COO)2·H2O
(0.1 mmol, 19.9 mg), with stirring. The mixture was stirred for a further 20 min at room temperature and then filtered.
The filtrate was kept in air for 5 d, during which time blue block-shaped crystals of (I) were formed.
S3. Refinement
The H atoms were placed in idealized positions and constrained to ride on their parent atoms, with C—H distances in the
Figure 1
The molecular structure of compound (I), showing the atom-numbering scheme. Displacement ellipsoids are drawn at the
supporting information
[image:5.610.159.459.70.518.2]sup-3
Acta Cryst. (2006). E62, m45–m46Figure 2
The crystal packing of compound (I), viewed along the a axis.
{4-Bromo-2-[(2-diethylaminoethylimino)methyl]phenolato}thiocyanatocopper(II)
Crystal data
[Cu(C13H18BrN2O)(NCS)] Mr = 419.82
Monoclinic, P21/c Hall symbol: -P 2ybc
a = 7.052 (1) Å
b = 16.688 (2) Å
c = 13.775 (2) Å
β = 94.79 (1)°
V = 1615.4 (4) Å3 Z = 4
F(000) = 844
Dx = 1.726 Mg m−3
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 3328 reflections
θ = 2.4–24.6°
µ = 3.96 mm−1 T = 298 K Block, blue
Bruker SMART CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
ω scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin = 0.404, Tmax = 0.510
11427 measured reflections 3564 independent reflections 2787 reflections with I > 2σ(I)
Rint = 0.031
θmax = 27.5°, θmin = 1.9° h = −8→9
k = −21→21
l = −16→17
Refinement
Refinement on F2 Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.036 wR(F2) = 0.091 S = 1.03 3564 reflections 192 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained
w = 1/[σ2(F
o2) + (0.0453P)2 + 0.4325P] where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001 Δρmax = 0.54 e Å−3 Δρmin = −0.41 e Å−3
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
Cu1 0.24192 (5) 0.72498 (2) 1.01867 (3) 0.03673 (12)
Br1 0.93758 (5) 0.40802 (2) 1.16694 (3) 0.05364 (13)
S1 −0.29725 (14) 0.82260 (8) 1.15452 (8) 0.0719 (3)
O1 0.2750 (3) 0.63306 (12) 1.09990 (15) 0.0410 (5)
N1 0.4757 (4) 0.70606 (14) 0.95910 (17) 0.0352 (5)
N2 0.2110 (4) 0.82183 (15) 0.92092 (18) 0.0390 (6)
N3 0.0115 (4) 0.75086 (19) 1.0795 (2) 0.0588 (8)
C1 0.5849 (4) 0.59285 (16) 1.0572 (2) 0.0324 (6)
C2 0.4214 (4) 0.58535 (16) 1.1096 (2) 0.0323 (6)
C3 0.4193 (4) 0.52132 (18) 1.1767 (2) 0.0375 (7)
H3 0.3135 0.5145 1.2119 0.045*
C4 0.5687 (4) 0.46902 (17) 1.1913 (2) 0.0382 (7)
H4 0.5629 0.4271 1.2355 0.046*
C5 0.7289 (4) 0.47863 (17) 1.1403 (2) 0.0359 (7)
C6 0.7373 (4) 0.53849 (17) 1.0741 (2) 0.0358 (6)
supporting information
sup-5
Acta Cryst. (2006). E62, m45–m46C7 0.6001 (4) 0.65218 (17) 0.9838 (2) 0.0348 (6)
H7 0.7099 0.6517 0.9508 0.042*
C8 0.5146 (5) 0.7638 (2) 0.8834 (3) 0.0514 (9)
H8A 0.5804 0.7376 0.8330 0.062*
H8B 0.5946 0.8067 0.9111 0.062*
C9 0.3304 (5) 0.7965 (2) 0.8414 (2) 0.0501 (8)
H9A 0.2633 0.7561 0.8013 0.060*
H9B 0.3531 0.8422 0.8004 0.060*
C10 0.2880 (6) 0.8978 (2) 0.9632 (3) 0.0696 (12)
H10A 0.4241 0.8914 0.9776 0.083*
H10B 0.2692 0.9392 0.9139 0.083*
C11 0.2081 (8) 0.9266 (3) 1.0516 (4) 0.1023 (18)
H11A 0.0741 0.9358 1.0382 0.153*
H11B 0.2695 0.9757 1.0724 0.153*
H11C 0.2283 0.8871 1.1021 0.153*
C12 0.0093 (5) 0.8275 (2) 0.8795 (3) 0.0513 (9)
H12A −0.0680 0.8425 0.9316 0.062*
H12B −0.0314 0.7745 0.8575 0.062*
C13 −0.0328 (5) 0.8853 (2) 0.7963 (3) 0.0573 (10)
H13A −0.0020 0.9388 0.8179 0.086*
H13B −0.1655 0.8825 0.7743 0.086*
H13C 0.0421 0.8714 0.7437 0.086*
C14 −0.1159 (5) 0.7801 (2) 1.1115 (2) 0.0465 (8)
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
Cu1 0.0362 (2) 0.0425 (2) 0.0329 (2) 0.00864 (15) 0.01138 (15) 0.00463 (15)
Br1 0.0387 (2) 0.0456 (2) 0.0759 (3) 0.00827 (14) 0.00099 (17) 0.00703 (16)
S1 0.0446 (5) 0.1075 (9) 0.0651 (7) 0.0174 (5) 0.0135 (5) −0.0313 (6)
O1 0.0387 (12) 0.0458 (12) 0.0412 (12) 0.0094 (9) 0.0189 (10) 0.0113 (9)
N1 0.0358 (13) 0.0378 (12) 0.0333 (13) 0.0006 (10) 0.0106 (11) 0.0022 (10)
N2 0.0379 (14) 0.0433 (14) 0.0358 (14) 0.0047 (11) 0.0037 (11) 0.0045 (11)
N3 0.0573 (19) 0.0691 (19) 0.0540 (19) 0.0265 (16) 0.0284 (15) 0.0197 (16)
C1 0.0330 (15) 0.0321 (14) 0.0323 (15) 0.0012 (11) 0.0030 (12) −0.0046 (12)
C2 0.0326 (15) 0.0356 (14) 0.0291 (15) −0.0005 (12) 0.0052 (12) −0.0038 (12)
C3 0.0373 (16) 0.0409 (16) 0.0354 (16) −0.0037 (13) 0.0091 (13) 0.0000 (13)
C4 0.0409 (17) 0.0346 (15) 0.0389 (17) −0.0021 (13) 0.0018 (14) 0.0010 (12)
C5 0.0295 (15) 0.0360 (15) 0.0414 (17) 0.0037 (12) −0.0026 (13) −0.0063 (13)
C6 0.0284 (14) 0.0397 (15) 0.0400 (17) −0.0002 (12) 0.0065 (12) −0.0064 (13)
C7 0.0291 (14) 0.0386 (15) 0.0383 (16) −0.0027 (12) 0.0115 (12) −0.0039 (12)
C8 0.052 (2) 0.0501 (19) 0.056 (2) 0.0058 (16) 0.0244 (17) 0.0156 (16)
C9 0.050 (2) 0.060 (2) 0.0425 (19) 0.0082 (16) 0.0150 (16) 0.0133 (16)
C10 0.068 (3) 0.064 (2) 0.073 (3) 0.007 (2) −0.011 (2) −0.009 (2)
C11 0.127 (5) 0.089 (3) 0.095 (4) 0.002 (3) 0.032 (4) −0.037 (3)
C12 0.0409 (19) 0.060 (2) 0.053 (2) 0.0056 (16) 0.0010 (16) 0.0171 (17)
C13 0.049 (2) 0.074 (2) 0.048 (2) 0.0158 (18) 0.0028 (17) 0.0182 (18)
Cu1—O1 1.902 (2) C5—C6 1.357 (4)
Cu1—N1 1.928 (3) C6—H6 0.9300
Cu1—N3 1.938 (3) C7—H7 0.9300
Cu1—N2 2.103 (2) C8—C9 1.482 (5)
Br1—C5 1.897 (3) C8—H8A 0.9700
S1—C14 1.617 (4) C8—H8B 0.9700
O1—C2 1.302 (3) C9—H9A 0.9700
N1—C7 1.282 (4) C9—H9B 0.9700
N1—C8 1.463 (4) C10—C11 1.464 (6)
N2—C10 1.480 (5) C10—H10A 0.9700
N2—C12 1.490 (4) C10—H10B 0.9700
N2—C9 1.497 (4) C11—H11A 0.9600
N3—C14 1.143 (4) C11—H11B 0.9600
C1—C6 1.411 (4) C11—H11C 0.9600
C1—C2 1.416 (4) C12—C13 1.509 (4)
C1—C7 1.426 (4) C12—H12A 0.9700
C2—C3 1.414 (4) C12—H12B 0.9700
C3—C4 1.370 (4) C13—H13A 0.9600
C3—H3 0.9300 C13—H13B 0.9600
C4—C5 1.389 (4) C13—H13C 0.9600
C4—H4 0.9300
O1—Cu1—N1 92.93 (9) N1—C8—C9 108.1 (3)
O1—Cu1—N3 89.52 (11) N1—C8—H8A 110.1
N1—Cu1—N3 176.48 (12) C9—C8—H8A 110.1
O1—Cu1—N2 176.09 (10) N1—C8—H8B 110.1
N1—Cu1—N2 84.27 (10) C9—C8—H8B 110.1
N3—Cu1—N2 93.41 (11) H8A—C8—H8B 108.4
C2—O1—Cu1 127.65 (19) C8—C9—N2 110.3 (3)
C7—N1—C8 119.1 (3) C8—C9—H9A 109.6
C7—N1—Cu1 126.3 (2) N2—C9—H9A 109.6
C8—N1—Cu1 114.43 (19) C8—C9—H9B 109.6
C10—N2—C12 113.8 (3) N2—C9—H9B 109.6
C10—N2—C9 108.6 (3) H9A—C9—H9B 108.1
C12—N2—C9 108.3 (3) C11—C10—N2 117.0 (4)
C10—N2—Cu1 113.0 (2) C11—C10—H10A 108.0
C12—N2—Cu1 109.78 (19) N2—C10—H10A 108.0
C9—N2—Cu1 102.55 (18) C11—C10—H10B 108.0
C14—N3—Cu1 167.6 (3) N2—C10—H10B 108.0
C6—C1—C2 120.0 (3) H10A—C10—H10B 107.3
C6—C1—C7 117.5 (3) C10—C11—H11A 109.5
C2—C1—C7 122.4 (3) C10—C11—H11B 109.5
O1—C2—C3 118.5 (3) H11A—C11—H11B 109.5
O1—C2—C1 124.5 (3) C10—C11—H11C 109.5
C3—C2—C1 117.0 (3) H11A—C11—H11C 109.5
supporting information
sup-7
Acta Cryst. (2006). E62, m45–m46C4—C3—H3 119.1 N2—C12—C13 116.9 (3)
C2—C3—H3 119.1 N2—C12—H12A 108.1
C3—C4—C5 120.0 (3) C13—C12—H12A 108.1
C3—C4—H4 120.0 N2—C12—H12B 108.1
C5—C4—H4 120.0 C13—C12—H12B 108.1
C6—C5—C4 120.5 (3) H12A—C12—H12B 107.3
C6—C5—Br1 120.7 (2) C12—C13—H13A 109.5
C4—C5—Br1 118.7 (2) C12—C13—H13B 109.5
C5—C6—C1 120.6 (3) H13A—C13—H13B 109.5
C5—C6—H6 119.7 C12—C13—H13C 109.5
C1—C6—H6 119.7 H13A—C13—H13C 109.5
N1—C7—C1 125.8 (3) H13B—C13—H13C 109.5
N1—C7—H7 117.1 N3—C14—S1 178.6 (3)