0095-1137/05/$08.00
⫹
0
doi:10.1128/JCM.43.6.2635–2641.2005
Copyright © 2005, American Society for Microbiology. All Rights Reserved.
High Frequency of Gastric Colonization with Multiple
Helicobacter pylori
Strains in Venezuelan Subjects
C. Ghose,
1G. I. Perez-Perez,
1* L. J. van Doorn,
2M. G. Domı´nguez-Bello,
3and M. J. Blaser
1,4Departments of Microbiology and Medicine, New York University School of Medicine, New York, New York
1; Delft
Diagnostic Laboratories, Delft, The Netherlands
2; Laboratory of Gastrointestinal Physiology, IVIC, Caracas,
Venezuela
3; and Department of Veterans Affairs Medical Center, New York, New York
4Received 29 November 2004/Returned for modification 22 December 2004/Accepted 17 February 2005
Multiple
Helicobacter pylori
strains may colonize an individual host. Using enzyme-linked immunosorbent
assay and line probe assay (LiPA) techniques, we analyzed the prevalence of mixed
H. pylori
colonization in 127
subjects from Venezuela, a country of high
H. pylori
prevalence, from three regions representing different
population groups: the Andes (Merida), where Caucasian mestizos predominate, a major city near the coast
(Caracas), where Amerindian-Caucasian-African mestizos predominate, and an Amazonian community
(Puerto Ayacucho), where Amerindians predominate and mestizos reflect Amerindian and Caucasian ancestry.
Among 121
H. pylori
-positive persons, the prevalence of
cagA
-positive strains varied from 50% (Merida) to 86%
(Puerto Ayacucho) by LiPA. Rates of mixed colonization also varied, as assessed by LiPA of the
vacA s
(mean,
49%) and
m
(mean, 26%) regions. In total, 55% of the individuals had genotypic evidence of mixed colonization.
vacA s1c
, a marker of Amerindian (East Asian) origin, was present in all three populations, especially from
Puerto Ayacucho (86%). These results demonstrate the high prevalence of mixed colonization and indicate that
the
H. pylori
East Asian
vacA
genotype has survived in all three populations tested.
Helicobacter pylori
is a gram-negative microaerophilic
bacte-rium that persistently colonizes the gastric mucosa of human
hosts for decades or life (14). More than 50% of the world’s
population carries
H. pylori
, with proportions as high as 80% in
developing countries (27). Although most
H. pylori
-positive
persons are asymptomatic, the presence of
H. pylori
is
associ-ated with increased risk for the development of peptic ulcer
disease, gastric adenocarcinoma, and gastric lymphoma (6, 46,
50). Expression of disease is associated with particular host and
bacterial factors (16, 17, 40, 47).
Two
H. pylori
genes,
vacA
and
cagA
, have been especially
associated with the differences in disease risk (19, 20, 61). In
the Western world,
cagA
is present in
⬃
60% of strains and is
the marker for a 35- to 40-kb pathogenicity island that encodes
a type IV secretion system responsible for the translocation of
the CagA protein into host gastric epithelial cells (2, 48).
Translocation of the CagA protein into host cells changes
signal transduction pathways and induces proinflammatory
cy-tokine production (31).
In contrast, all
H. pylori
strains contain
vacA
, but its product
is detectable in vitro in only about half the strains (20). The
vacA
genotypes for any given
H. pylori
strain are a mosaic of
combinations of signal sequence and midregion genotypes or
are chimerae (9). The
vacA
signal sequence (s sequence) has
two major genotypes,
s1
and
s2
, and there are three variations
of
s1
:
s1a
,
s1b
, and
s1c
(62, 64). The midregion of
vacA
, about
700 bp, has two major genotypes,
m1
and
m2
(9, 10, 21). Strains
of
vacA s1
/
m1
and
s1
/
m2
types produce high and moderate
levels of vacuolating activity, respectively, whereas
s2
/
m2
strains produce little or none (9). Particular
vacA
genotypes
vary in their geographic prevalence and serve as markers for
the ancestry of the
H. pylori
isolates; for example,
vacA s1c
is a
strong marker for East Asian ancestry (24, 29, 62, 68).
H. pylori
possesses an unusually high number of type II
restriction-modification (R-M) systems, and each strain varies
in its complement of R-M systems (58, 69). For the
hpyI
and
the
hpyIII
R-M systems, the methyltransferase gene (either
hpyIM
or
hpyIIIM
) is present in all strains, but in some strains
hpyIR
has been replaced with another gene (
hrgB
) and/or
hpyIIIR
has been replaced by
hrgA
(5, 25). Each strain
pos-sesses either gene at the
hpyIR
locus and not both or neither;
the same has been found for the
hpyIIIR
locus (4). Therefore,
the detection of both alleles in a single gastric specimen
im-plies that the host is colonized by multiple
H. pylori
strains.
Because multiple
H. pylori
strains may colonize a single
patient, intergenomic recombination occurs, and the
H. pylori
population structure indicates that such events have been
rel-atively common (12, 38, 57). Colonization by multiple
H. pylori
strains appears more common in countries where
H. pylori
is
highly prevalent (8, 36, 43, 45).
Venezuela, a country of high
H. pylori
prevalence, was
set-tled by persons of different ethnicities, and ethnic mixing
con-tinues to the present (22, 44). Nevertheless, in the South
(Amazonas), Amerindians predominate and mestizos reflect
Amerindian and Caucasian ancestry; in the Western Andes
(Merida), Caucasian mestizos predominate; and in the
North-Central
region
(Caracas),
Amerindian-Caucasian-African
mestizos predominate (18, 52).
In this study, we analyzed the prevalence of mixed
H. pylori
colonization among persons in these three different locales in
Venezuela. We hypothesized that we would find evidence for
mixed colonization and that the circulating strains would
re-flect the ethnicities of the host population. Our analysis was
* Corresponding author. Mailing address: Infectious Diseases
Lab-oratories, Department of Medicine, VAMC 6026 West, 423 East 23rd
Street, New York, NY 10010. Phone: (212) 4105. Fax: (212)
263-4108. E-mail: perezg02@med.nyu.edu.
2635
on May 16, 2020 by guest
http://jcm.asm.org/
based on
vacA s
and
m
genotypes, using line probe assay
(LiPA), and on
cagA
prevalence, using enzyme-linked
immu-nosorbent assay (ELISA), and in one locale we used R-M
alleles to define the extent of mixed colonization. Using
biop-sy-based methods instead of pure isolated strain-based
meth-ods and using relatively simple and widely available marker
systems, we sought to determine the extent to which
cocoloni-zation with multiple
H. pylori
strains is present in the three
Venezuelan populations studied. We also sought to determine
whether the distribution of circulating
vacA
alleles is similar in
the three locales, thus reflecting the origins of the
H. pylori
strains (whether East Asian or Western).
MATERIALS AND METHODS
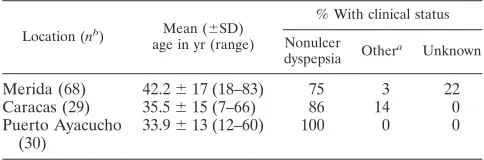
Subjects.DNA was obtained from antrum biopsies from patients from Merida in the Andean state, from Caracas, the urban capital of Venezuela, and from Puerto Ayacucho, where medical care is provided to populations from surround-ing Amazonian villages. The facilities at Puerto Ayacucho, Caracas, and Merida are parts of the public hospital system, where patients of lower socioeconomic status go for medical care. All of the studied patients were of lower socioeco-nomic status. The 30 persons studied from the Puerto Ayacucho area (mean age, 34 years), were of Amerindian ancestry (25 Piaroas and 5 Guajibos) and are representative of a rural poor community in the Amazonas state (Table 1). All 29 persons from Caracas (mean age 36, years) represent a poor urban community where Amerindian-Caucasian-African mestizos predominate. The 68 persons from Merida (mean age, 43 years) are representative of a poor urban community in the Andes where Caucasian mestizos predominate.
From each patient two antrum biopsies were taken, 2 cm from the pylorus, one for culture and one for PCR. All patients were undergoing upper gastrointestinal endoscopy for dyspeptic symptoms and signed a consent form to participate in this study. The prevalence of peptic ulcer disease is low in all three populations, as has been reported previously (22).
In our previous study, we analyzed the prevalence of thevacA s1callele in populations from Caracas and Puerto Ayacucho by PCR followed by sequencing (29). We were able to subject to PCR and sequence 13 samples from Caracas and 16 samples from Puerto Ayacucho at thevacA slocus, of which 7 (54%) and 12 (75%), respectively, are included in our current study.
Serum samples also were collected from the subjects in Caracas and Puerto Ayacucho but not from Merida. We were not able to establish ethnicity for individual patients.
Specimens for LiPA analysis.DNA was extracted from biopsies using the QIAGEN DNeasy tissue kit (Valencia, CA). One hundred nanograms of total genomic DNA was used for PCR. PCR was performed using biotin-labeled primers specific forvacA s,m, and cagA, as described previously (64). Ten microliters of each biotin-labeled PCR product was denatured by the addition of 10 ml of 400 mM NaOH and 10 mM EDTA and used in the subsequent reverse hybridization LiPA assay.
Reverse-hybridization LiPA analysis.PCR-LiPA was used to study the pres-ence ofcagAand the differentvacA sandmgenotypes in the patient specimens (64, 65). Briefly, allele-specific oligonucleotide probes forvacA s1a,s1b, ands1c,
vacA m1andm2, andcagAwere tailed with poly(dT) and were immobilized as parallel lines on nitrocellulose strips, as described previously (65). The LiPA strips were incubated in prewarmed hybridization solution, and then denatured
PCR products from the patient specimens were added. After 30 min of incuba-tion at room temperature, the strips were rinsed again, and 4-nitroblue tetrazo-lium chloride and 5-bromo-4-chloro-3-indolylphosphate substrate were added. Positive hybridizations were visible as purple probe lines. Results were read at the moment of experiments, since positive bands fade over time. Moreover, results are qualitative and not quantitative; hence, no assumptions can be made as to which allele is the major/minor type present in one sample. DNAs fromH. pyloristrains with known genotypes were used as positive and negative controls. The LiPA has been previously used for detectingH. pyloriin gastric biopsy samples, formalin-fixed samples, and paraffin-embedded samples and shows su-perior sensitivity compared with culture and histopathology (54, 55, 66). A sensitivity analysis was not performed for the LiPA, since it is a well-established technique with a detection capacity better than 1 in 20 (5%) to 1:10,000 (0.001%), as shown for hepatitis C virus and human papillomavirus detection (37, 67).
Whole-cell and CagA ELISA.The presence of immunoglobulin G (antibodies to anH. pyloriwhole-cell (WC) antigen preparation or to CagA protein was assessed by enzyme-linked immunosorbent assays (ELISA), as described previ-ously (28, 39). In brief, each antigen was dissolved in 0.5 M sodium bicarbonate (pH 9.6) and used to coat microtiter plates overnight at room temperature. Patient serum was added at 1:800 and 1:100 dilutions for WC and CagA anti-bodies, respectively, and incubated for 1 h at 37°C. Human anti-immunoglobulin G conjugated to horseradish peroxidase (Biosource International, Camarillo, CA) was used, and the mixture reincubated for 1 h. ABTS [2–2⬘-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid)] (Sigma Aldrich, St. Louis, MO) was added as a substrate, and plates were read at 405 nm. Each serum sample was tested in duplicate. Data were analyzed using the Revelation 2.0 program (Dynatech, Westwood, NJ). For WC, serum samples with ELISA units ofⱖ1.0 were con-sidered seropositive, while a value of⬍1 was considered negative. For CagA, serum samples with ELISA units ofⱖ0.35 were considered seropositive, while a value of⬍0.35 was considered negative (39).
Analysis of R-M systems.The presence of the restriction endonuclease or replacing gene in both thehpyIandhpyIIIR-M loci was analyzed using PCR in 31 samples from Merida. Analysis was limited to a subset of Merida biopsies in which there was sufficient residual DNA for these PCR amplifications. Primers and PCR conditions forhpyIR,hrgB,hpyIIIR, andhrgAhave been described previously (5, 25). The presence of bothhpyIRandhrgBPCR products orhpyIIIR
andhrgAPCR products in DNA from a single biopsy sample was considered evidence for the presence of multiple strains.
Sensitivity analysis of R-M system allele PCR.A sensitivity analysis of the PCR for each R-M system gene used in the detection of mixed colonization was performed using genomic DNA fromH. pyloristrains. For thehpyIRallele, genomic DNA from strain 26695 (hpyIR⫹hrgB⫺) was analyzed at concentra-tions from 100 ng per reaction to 0.001 ng per reaction as template DNA. For each reaction, genomic DNA from strain J99 (hrgB⫹hpyIR⫺) was added at 100 ng per reaction as competing DNA. The converse concentrations were used for the sensitivity analysis of thehrgBPCR. For thehpyIIIRallele, dilutions were made with genomic DNA from 26695 (hpyIIIR⫹hrgA⫺) as template DNA from 100 ng to 0.001 ng per reaction. Genomic DNA from strain 60190 (hrgA⫹
hpyIIIR⫺) was added at 100 ng per reaction as competing DNA. The converse concentrations were used for the sensitivity analysis of thehrgAallele. PCR conditions were as described previously (5, 25).
Statistical analysis.A chi-square test was used to compare the frequencies of mixed or single colonizations in patients from the different locales. APvalue of ⬍0.05 was considered to be significant.
RESULTS
Comparison of ELISA and LiPA methodologies for
H. pylori
detection and
cagA
prevalence in the studied populations. Of
the 127 subjects studied, 121 (95%) had evidence by LiPA for
H. pylori
carriage (Table 2). The prevalence of
cagA
-positive
[image:2.585.42.284.90.170.2]strains in these 121 subjects, as determined by LiPA, varied in
the three localities: Merida (50%), Caracas (77%), and Puerto
Ayacucho (86%) (
P
⬍
0.0001). The results of the LiPA analysis
of the strains and the ELISA analysis of the serum from the
patients were compared with samples from Caracas and Puerto
Ayacucho. For the specimens from Puerto Ayacucho, the two
methodologies provide nearly identical results. For the
speci-mens from Caracas, detection of carriage of
cagA
-positive
H.
TABLE 1. Characteristics of the 127 subjects in the 3 studied
populations in Venezuela
Location (nb) Mean (⫾SD) age in yr (range)
% With clinical status
Nonulcer dyspepsia Other
a
Unknown
Merida (68)
42.2
⫾
17 (18–83)
75
3
22
Caracas (29)
35.5
⫾
15 (7–66)
86
14
0
Puerto Ayacucho
(30)
33.9
⫾
13 (12–60)
100
0
0
aDuodenal ulcer (n⫽5), Barrett’s esophagus (n⫽1). bNo. of subjects.
on May 16, 2020 by guest
http://jcm.asm.org/
pylori
strains was observed in 77% and 61% of the individuals,
using the LiPA and ELISA methodologies, respectively.
Be-tween the two populations, using the LiPA as the “gold
stan-dard,” the CagA ELISA had five false negatives (9%) and two
false positives (14%). In total, these studies showed that
H.
pylori
is highly prevalent in these populations, with a mixture of
cagA
-positive and
cagA-
negative strains.
Prevalence of specific
vacA s
genotypes in the three studied
populations of Venezuela.
LiPA analysis of DNA extracted
from biopsies can detect colonization by
H. pylori
strains that
differ in
vacA s
and
vacA m
genotypes (26). Since
H. pylori
cells
possess only a single copy of
vacA
, the presence of more than
one
vacA s
or
m
genotype in a DNA sample indicates
coloni-zation by multiple strains (26).
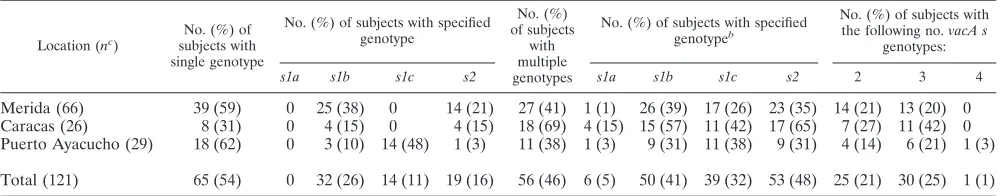
Of the 121 biopsies studied, a single
vacA s
genotype was
observed in 65 (54%) of the samples, with the prevalence
ranging from 31% (Caracas) to 62% (Puerto Ayacucho)
(Ta-ble 3). Conversely, multiple genotypes were observed in 69%,
38%, and 41% of the specimens from Caracas, Puerto
Ayacu-cho, and Merida, respectively. In the 56 biopsies (46%) from
the three populations combined that showed multiple
vacA s
genotypes, 148 (mean, 2.57
⫾
0.53 per biopsy) alleles were
observed, with frequencies varying between 2.48
⫾
0.50 (in
Merida) and 2.72
⫾
0.65 (in Puerto Ayacucho). In Caracas, the
most common circulating genotype was
vacA s2
, appearing in
21 (81%) of the subjects. In Merida,
vacA s1b
occurred in 51
(77%), and in Puerto Ayacucho,
vacA s1c
occurred in 25 (86%)
of the subjects. The
vacA s1a
genotype never was detected as
a single genotype in any patient, whereas
vacA s1c
was
de-tected as a single genotype only in Puerto Ayacucho (48%).
Twenty-five (86%) of the specimens from Puerto Ayacucho
contained
vacA s1c
, a significantly greater proportion than in
Merida (26%;
P
⬍
0.0001) and in Caracas (42%;
P
⫽
0.001).
Prevalence of specific
vacA m
genotypes in the three studied
populations of Venezuela.
Only two
vacA m
genotypes were
distinguishable by LiPA (Table 4). Eleven percent of the
spec-imens examined by the
vacA m
LiPA yielded untypable results,
versus none for the
vacA s
genotype LiPA, reflecting the
greater genetic complexity of the
vacA m
locus (10). In total, 24
(20%) of the 121 specimens analyzed showed evidence for both
vacA m1
and
m2
genotypes (Table 4). Mixed colonization was
detected most often in Puerto Ayacucho (52%) and least in
Merida (6%). The
vacA m1
genotype was present in 90% of
specimens from Puerto Ayacucho, ranging to 46% in Caracas.
The
vacA m2
genotype ranged from 62% in Puerto Ayacucho
to 36% in Merida. In total, based on multiple genotypes in
either the
vacA s
or
m
loci, the prevalence of mixed
coloniza-tion detected in the specimens was 55% (Table 4).
This methodology could permit detection of up to four
vacA
s
genotypes and two
vacA m
genotypes. In Caracas, the
ma-jority of persons with mixed colonization carried three
vacA s
alleles (42%), whereas in Merida and Puerto Ayacucho, the
majority were colonized with a single
vacA s
genotype (59%
and 62%, respectively). In Puerto Ayacucho, there was
evi-dence in one subject for all four
vacA s
genotypes (Table 3).
[image:3.585.43.545.90.177.2]Sensitivity analysis of R-M alleles as a tool for detection of
mixed colonization.
For the
hpyIR
and
hpyIIIR
alleles, the
primers were able to amplify a product from 0.01 ng of
tem-plate DNA when the majority of the DNA present in the PCR
mix contained genomic DNA from strains that carried the
heterologous allele (Fig. 1). For the
hrgA
and
hrgB
alleles, the
TABLE 2.
H. pylori
and
cagA
status for 127 subjects from 3 locations in Venezuela as determined from analysis of gastric specimens by LiPA
and serum IgG antibody responses by ELISA
Patient status (H. pylori,
cagA)
No. (%) of subjects in category
Merida (n⫽68) Caracas (n⫽29) Puerto Ayacucho (n⫽30) Total (n⫽127)
LiPA ELISA LiPA ELISA LiPA ELISA LiPA
⫺
,
⫺
2 (4)
ND
a3 (10)
0
1 (3)
0
6 (5)
⫹
,
⫺
33 (48)
ND
6 (21)
13 (45)
4 (13)
4 (13)
43 (34)
⫺
,
⫹
0
ND
0
0
0
2 (7)
0 (0)
⫹
,
⫹
33 (48)
ND
20 (69)
16 (55)
25 (83)
24 (80)
78 (61)
aND, not determined, because serum specimens were not available from patients in Merida.
TABLE 3. Distribution of
vacA s
genotypes in gastric samples from 121
H. pylori-positive subjects in 3 studied populations in Venezuela
aLocation (nc )
No. (%) of subjects with single genotype
No. (%) of subjects with specified genotype
No. (%) of subjects
with multiple genotypes
No. (%) of subjects with specified genotypeb
No. (%) of subjects with the following no.vacA s
genotypes:
s1a s1b s1c s2 s1a s1b s1c s2 2 3 4
Merida (66)
39 (59)
0
25 (38)
0
14 (21)
27 (41)
1 (1)
26 (39)
17 (26)
23 (35)
14 (21)
13 (20)
0
Caracas (26)
8 (31)
0
4 (15)
0
4 (15)
18 (69)
4 (15)
15 (57)
11 (42)
17 (65)
7 (27)
11 (42)
0
Puerto Ayacucho (29)
18 (62)
0
3 (10)
14 (48)
1 (3)
11 (38)
1 (3)
9 (31)
11 (38)
9 (31)
4 (14)
6 (21)
1 (3)
Total (121)
65 (54)
0
32 (26)
14 (11)
19 (16)
56 (46)
6 (5)
50 (41)
39 (32)
53 (48)
25 (21)
30 (25)
1 (1)
aExcludes the six persons whose biopsy did not yield anH. pylorisignal. bPercentages total⬎100, since there were multiple genotypes per subject. cNo. of subjects.
on May 16, 2020 by guest
http://jcm.asm.org/
[image:3.585.43.543.601.699.2]primers were able to amplify a product from 0.001 ng of
tem-plate DNA.
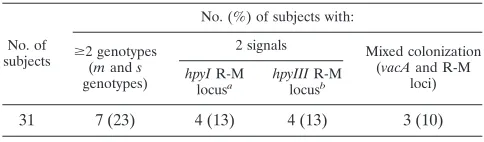
Analysis of mixed colonization using R-M system genes.
Of
the 68 samples from Merida, 31 samples were further analyzed
for the prevalence of mixed colonization based on the
hpyIR
and
hpyIIIR
loci (Fig. 2). Of the 31 samples analyzed, 8 (26%)
showed the presence of mixed colonization, based on results
involving either the
hpyIII
or
hpyI
locus (Table 5). For three
(38%) of these eight samples, mixed colonization also had
been detected by LiPA at the
vacA s
or
m
loci; thus, five new
samples of mixed colonization were detected.
DISCUSSION
Studies comparing
vacA
sequences in isolates from different
countries found geographic variation in the distribution of
genotypes
s1a
,
s1b
, and
s1c
and differences in
m1
and
m2
distribution (10, 62). As elsewhere, the
H. pylori
alleles
cur-rently present in Venezuelans reflect the ancestry of their
[image:4.585.44.542.80.185.2]progenitors (24, 29). That the population in Puerto Ayacucho,
predominantly Amerindian, carries
H. pylori
with high
preva-lences of
cagA
and
vacA s1c
genotypes, as in East Asian
com-munities, extends previous analyses suggesting a link between
ancestral South American and East Asian populations (24, 29,
70). Although the
s1c
genotype was found in fewer than half of
the study subjects in the two other regions, its presence is an
indication of persistence despite the admixture of European
and African strains, as competition and recombination
contin-ues (23, 24). The mixtures of strains found in the host
popu-lations reflect the mixing of the human ethnicities introduced
into Latin America since Columbus. In the “less mestiza”
Puerto Ayacucho, the ancestral marker (
vacA s1c
) was found
to be much less diluted with European (
vacA s1b
or
vacA s2
)
strains than in Merida and Caracas. No other study has
previ-ously reported the presence of
H. pylori
strains possessing the
East Asian marker (
vacA s1c
) in major cities in Latin America
(35, 70). The prevalence of
vacA s1c
in 86% of the specimens
[image:4.585.115.473.469.684.2]FIG. 1. Sensitivity analysis of R-M system alleles for detecting mixed colonization using genomic DNA. For the positive control, the template
DNA (T) is 100 ng of homologous genomic DNA. For one negative control, competing heterologous DNA (C) was used at 100 ng. For each
reaction, template DNA at 100 ng, 10 ng, 1 ng, 0.1 ng, 0.01 ng, or 0.001 ng was added to 100 ng of competing DNA. Another negative control
included water (W) only.
TABLE 4. Distribution of
vacA m
genotypes in gastric samples from 121
H. pylori-positive subjects in 3 studied populations in Venezuela
aLocation (nd)
mgenotypes
mandsgenotypes: No. (%) of subjects withⱖ2
genotypesc No. (%) of subjects
with single genotype
No. (%) with
specified genotype No. (%) of subjects withboth m1 and m2 genotypes
No. (%) of subjects with untypable
samplesb
m1 m2
Merida (66)
51 (77)
31 (47)
20 (30)
4 (6)
11 (16)
27 (41)
Caracas (26)
19 (73)
7 (27)
12 (46)
5 (19)
2 (8)
20 (77)
Puerto Ayacucho (29)
14 (48)
11 (38)
3 (10)
15 (52)
0
19 (66)
Total (121)
84 (69)
49 (40)
35 (29)
24 (20)
13 (11)
66 (55)
aExcludes the six persons whose biopsy did not yield anH. pylorisignal. bPercentages total⬎100, since there were multiple genotypes per subject.
cIncluding diversity of bothvacA s-genotypes (see Table 3) andm-genotypes in the same individual. dNo. of subjects.
on May 16, 2020 by guest
http://jcm.asm.org/
from Puerto Ayacucho, present either as a single or as a mixed
colonization, is significantly higher than in either Merida or
Caracas; in the latter cities,
vacA s1c
only was present in mixed
colonization. We were able to identify the minor allele
popu-lations in Caracas and Merida because we used the LiPA assay.
LiPA is a more-sensitive method for the detection of mixed
infection than is sequencing, with capability of detecting minor
populations at one part in 20 (5%) to up to one part in 10,000
(0.001%) (37, 54, 55). With PCR and sequencing, usually only
the major allele is detected, and thus, minor populations were
not well appreciated in previous work (29, 63, 66).
The detection in a gastric specimen of multiple alleles at loci
in which only one allele can be present per strain implies that
the host is harboring multiple different isolates. The simplest
interpretation is that the multiple isolates were present in the
specimen at the time of sampling. An alternative explanation is
that multiple strains once were present in that host and that
recombination occurred before one (or more) of the strains
was lost. Thus, the remaining strain(s) represents mixed
pop-ulations that are largely similar but vary at the particular
al-leles. Recent data provide strong support that this
phenome-non occurs as well (23). Regardless of whether the differences
observed reflect recombination or the actual presence of two
entirely distinct strains, detection of multiple alleles indicates
the presence of multiple strains in a host, past or current.
In this study, evidence of colonization with multiple
H. pylori
strains was frequently detected. Our results likely
underesti-mate the extent of mixed colonization, particularly in places
like Puerto Ayacucho, where the circulating strains are less
diverse; mixed colonization with strains of the same
vacA
ge-notypes could not be detected in our analysis. Analysis of
additional multiallelic genes would allow the identification of a
greater extent of mixed infection. Analysis of biallelic loci in
two R-M systems provided independent confirmation of the
general phenomenon of colonization by multiple
H. pylori
strains. For example, in Merida, of 31 samples analyzed for
R-M system alleles, 5 samples that were previously believed to
show a single colonization based on
vacA s
and
m
LiPA were
identified as having mixed colonization based on R-M system
alleles. Moreover, analysis of more than one biopsy per patient
from more than one location (antrum and corpus) would
in-crease the power of detection of mixed colonization, as has
been reported previously (30).
Our major conclusions are that colonization by multiple
different
H. pylori
strains is more common than previously
reported and that the greater the number of polymorphic
markers used, the greater the extent of the intrahost diversity
appreciated. Previous studies have not reported or have
un-derreported mixed colonization due to the techniques that
have been used, chiefly culture-based methods followed by
sequencing. The use of high-resolution techniques such as
LiPA and the analysis of multiple loci have led to the
under-standing that mixed colonization is highly prevalent.
van Doorn et al. found an 11% rate of multiple strains in
hosts from northern South America, respectively (62). Other
reports of colonization with multiple strains from different
geographic locales have included Korea (60%), Brazil (15%),
Mexico (65%), Chile (32%), and Portugal (30%) (8, 26, 36, 45,
63). In a study of 400 biopsy samples from 20
H. pylori
-positive
subjects in Mexico, there was evidence for colonization with
multiple strains in 17 (85%) of the 20 subjects (45). Population
genetics studies of
H. pylori
indicate a history of substantial
recombination; the high rates of mixed colonization that we
and others have detected provide the substrate for this
exten-sive recombination (1, 7, 23, 57).
H. pylori
has been disappearing with socioeconomic
[image:5.585.136.452.69.195.2]devel-opment (11, 49, 51, 53). As hygienic conditions have improved,
leading to diminishing
H. pylori
transmission, the mean
num-ber of strains per colonized person likely has declined, a
phe-nomenon implying that the overall extinction of
H. pylori
may
be greater than previously appreciated (13). If loss of
recom-bination possibilities affects the health of the
H. pylori
popu-lation in an individual host, the likelihood of persistence, and
FIG. 2. Agarose gel electrophoresis of PCR products involving the
hpyI
(locus 1) and
hpyIII
(locus 2) R-M loci in gastric biopsies from patients
M20 and M67. Lanes indicate PCR products or their absence at each locus. Primers amplified
hpyIR
and/or
hrgB
at locus 1 and
hpyIIIR
and/or
hrgA
at locus 2. In both patients, there is evidence for at least two different strains in each gastric biopsy.
TABLE 5. Distribution of
vacA m
and
s
genotypes and R-M system
genes in gastric samples from 31
H. pylori-positive subjects in
Merida, Venezuela
No. of subjects
No. (%) of subjects with:
ⱖ2 genotypes (mands
genotypes)
2 signals Mixed colonization (vacAand R-M
loci)
hpyIR-M
locusa hpyIIIR-M locusb
31
7 (23)
4 (13)
4 (13)
3 (10)
ahpyIRandhrgB. bhpyIIIRandhrgA.
on May 16, 2020 by guest
http://jcm.asm.org/
[image:5.585.42.284.639.710.2]thus transmission, may have been diminishing, further
accel-erating
H. pylori
’s disappearance.
Evidence in animal models suggests that colonization with
one particular “founding” strain may diminish subsequent
col-onization by another strain (32, 33, 41, 42, 56, 59, 60). The
likelihood of establishing new colonization may depend upon
whether the new strain has a selective advantage (15). The
presence of coexisting strains of
Helicobacter pylori
within a
single patient may be explained by limited competition for the
same mucosal surface by competing strains (34). The evidence
of mixed colonization can be utilized in mathematical models
to better define parameter values; however, whether particular
H. pylori
genotypes confer survival advantages may be both
host and context specific (3, 15).
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grant
R01GM63270, the Medical Research Service of the Department of
Veterans Affairs, the Institute for Urban and Global Health (NYU),
the Instituto Venezolano de Investigaciones Cientificas, and the
Filomena D’Agostino Foundation.
We acknowledge the collaboration of the Gastroenterology Service
at Hospital General del Oeste in Caracas, Euclides Gonzalez from the
Hospital Central de Puerto Ayacucho, Hospital Jose Gregorio
Her-nandez in Puerto Ayacucho, and Imad Kansau at the Universidad de
Los Andes in Merida.
REFERENCES
1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn.1999. Recombination and clonal groupings withinHelicobacter pylorifrom differ-ent geographical regions. Mol. Microbiol.32:459–470.
2.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg.1998. Analyses of the cag pathogenicity island ofHelicobacter pylori. Mol. Micro-biol.28:37–53.
3.Akopyants, N. S., K. A. Eaton, and D. E. Berg.1995. Adaptive mutation and cocolonization duringHelicobacter pyloriinfection of gnotobiotic piglets. Infect. Immun.63:116–121.
4.Ando, T., R. A. Aras, K. Kusugami, M. J. Blaser, and T. M. Wassenaar.2003. Evolutionary history ofhrgA, which replaces the restriction genehpyIIIRin thehpyIIIlocus ofHelicobacter pylori. J. Bacteriol.185:295–301.
5.Ando, T., T. M. Wassenaar, R. M. Peek, Jr., R. A. Aras, A. I. Tschumi, L. J. van Doorn, K. Kusugami, and M. J. Blaser. 2002. AHelicobacter pylori
restriction endonuclease-replacing gene,hrgA, is associated with gastric can-cer in Asian strains. Cancan-cer Res.62:2385–2389.
6.Anonymous.1994. Infection withHelicobacter pylori. IARC Monogr. Eval. Carcinog. Risks Hum.61:117–240.
7.Aras, R. A., J. Kang, A. I. Tschumi, Y. Harasaki, and M. J. Blaser.2003. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc. Natl. Acad. Sci. USA100:13579–13584.
8.Ashour, A. A., P. P. Magalhaes, E. N. Mendes, G. B. Collares, V. R. de Gusmao, D. M. Queiroz, A. M. Nogueira, G. A. Rocha, and C. A. de Oliveira.
2002. Distribution ofvacAgenotypes inHelicobacter pyloristrains isolated from Brazilian adult patients with gastritis, duodenal ulcer or gastric carci-noma. FEMS Immunol. Med. Microbiol.33:173–178.
9.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover.1995. Mosaicism in vacuolating cytotoxin alleles ofHelicobacter pylori. Association of specificvacA types with cytotoxin production and peptic ulceration. J. Biol. Chem.270:17771–17777.
10.Atherton, J. C., P. M. Sharp, T. L. Cover, G. Gonzalez-Valencia, R. M. Peek, Jr., S. A. Thompson, C. J. Hawkey, and M. J. Blaser.1999. Vacuolating cytotoxin (vacA) alleles ofHelicobacter pyloricomprise two geographically widespread types, m1 and m2, and have evolved through limited recombi-nation. Curr. Microbiol.39:211–218.
11.Banatvala, N., K. Mayo, F. Megraud, R. Jennings, J. J. Deeks, and R. A. Feldman.1993. The cohort effect andHelicobacter pylori. J. Infect. Dis.
168:219–221.
12.Beji, A., P. Vincent, I. Darchis, M. O. Husson, A. Cortot, and H. Leclerc.
1989. Evidence of gastritis with severalHelicobacter pyloristrains. Lancet
2:1402–1403.
13.Blaser, M. J.1998. Helicobacters are indigenous to the human stomach: duodenal ulceration is due to changes in gastric microecology in the modern era. Gut43:721–727.
14.Blaser, M. J., and J. C. Atherton. 2004.Helicobacter pyloripersistence: biology and disease. J. Clin. Investig.113:321–333.
15.Blaser, M. J., and D. Kirschner. 1999. Dynamics of Helicobacter pylori
colonization in relation to the host response. Proc. Natl. Acad. Sci. USA
96:8359–8364.
16.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura.1995. Infection with
Helicobacter pyloristrains possessingcagAis associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res.55:2111–2115. 17.Carroll, I. M., A. A. Khan, and N. Ahmed.2004. Revisiting the pestilence of
Helicobacter pylori: insights into geographical genomics and pathogen evo-lution. Infect. Genet. Evol.4:81–90.
18.Castro de Guerra, D., H. Arvelo, and J. Pinto-Cisternas.1999. Population structure of two black Venezuelan populations studied through their mating structure and other related variables. Ann. Hum. Biol.26:141–150. 19.Cover, T. L.1996. The vacuolating cytotoxin ofHelicobacter pylori. Mol.
Microbiol.20:241–246.
20.Cover, T. L., and M. J. Blaser.1992. Purification and characterization of the vacuolating toxin fromHelicobacter pylori. J. Biol. Chem.267:10570–10575. 21.Cover, T. L., M. K. Tummuru, P. Cao, S. A. Thompson, and M. J. Blaser.
1994. Divergence of genetic sequences for the vacuolating cytotoxin among
Helicobacter pyloristrains. J. Biol. Chem.269:10566–10573.
22.Dominguez-Bello, M. G., B. Beker, M. Guelrud, J. Vivas, S. Peraza, M. E. Perez, and L. R. Pericchi.2002. Short report: socioeconomic and seasonal variations ofHelicobacter pyloriinfection in patients in Venezuela. Am. J. Trop. Med. Hyg.66:49–51.
23.Falush, D., C. Kraft, N. S. Taylor, P. Correa, J. G. Fox, M. Achtman, and S. Suerbaum.2001. Recombination and mutation during long-term gastric col-onization byHelicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA98:15056–15061.
24.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Me-graud, K. Otto, U. Reichard, E. Katzowitsch, X. Wang, M. Achtman, and S. Suerbaum.2003. Traces of human migrations inHelicobacter pylori popula-tions. Science299:1582–1585.
25.Figueiredo, C., W. G. Quint, R. Sanna, E. Sablon, J. P. Donahue, Q. Xu, G. G. Miller, R. M. Peek, Jr., M. J. Blaser, and L. J. van Doorn.2000. Genetic organization and heterogeneity of theiceAlocus ofHelicobacter pylori. Gene246:59–68.
26.Figueiredo, C., L. J. Van Doorn, C. Nogueira, J. M. Soares, C. Pinho, P. Figueira, W. G. Quint, and F. Carneiro.2001.Helicobacter pylorigenotypes are associated with clinical outcome in Portuguese patients and show a high prevalence of infections with multiple strains. Scand. J. Gastroenterol.36:
128–135.
27.Frenck, R. W., Jr., and J. Clemens.2003. Helicobacter in the developing world. Microbes Infect.5:705–713.
28.Garza-Gonzalez, E., F. J. Bosques-Padilla, R. Tijerina-Menchaca, J. P. Flores-Gutierrez, H. J. Maldonado-Garza, and G. I. Perez-Perez. 2003. Comparision of endoscopy-based and serum-based methods for the diagno-sis ofHelicobacter pylori. Can. J. Gastroenterol.17:101–106.
29.Ghose, C., G. I. Perez-Perez, M. G. Dominguez-Bello, D. T. Pride, C. M. Bravi, and M. J. Blaser.2002. East Asian genotypes ofHelicobacter pylori
strains in Amerindians provide evidence for its ancient human carriage. Proc. Natl. Acad. Sci. USA99:15107–15111.
30.Hennig, E. E., L. Trzeciak, J. Regula, E. Butruk, and J. Ostrowski.1999. VacA genotyping directly from gastric biopsy specimens and estimation of mixed Helicobacter pyloriinfections in patients with duodenal ulcer and gastritis. Scand. J. Gastroenterol.34:743–749.
31.Higashi, H., R. Tsutsumi, S. Muto, T. Sugiyama, T. Azuma, M. Asaka, and M. Hatakeyama.2002. SHP-2 tyrosine phosphatase as an intracellular target ofHelicobacter pyloriCagA protein. Science295:683–686.
32.Hua, J., K. L. Ling, H. S. Ng, and B. Ho.2000. Isolation of a single strain of
Helicobacter pylorifrom the antrum and body of individual patients. Eur. J. Gastroenterol. Hepatol.12:1129–1134.
33.Hua, J., H. C. Ng, K. G. Yeoh, and B. Ho.1999. Predominance of a single strain ofHelicobacter pyloriin gastric antrum. Helicobacter4:28–32. 34.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow,
and R. M. Peek, Jr.2001.Helicobacter pylorigenetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA98:14625– 14630.
35.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. Su, Z. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcon, M. Lopez-Brea, G. Balakrish Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Wong, S. K. Lam, F. O. Olfat, T. Boren, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg.2000. Differences in genotypes ofHelicobacter pylorifrom different human populations. J. Bacteriol.182:3210–3218.
36.Kim, S. Y., C. W. Woo, Y. M. Lee, B. R. Son, J. W. Kim, H. B. Chae, S. J. Youn, and S. M. Park.2001. Genotyping CagA, VacA subtype, IceA1, and BabA ofHelicobacter pyloriisolates from Korean patients, and their associ-ation with gastroduodenal diseases. J. Korean Med. Sci.16:579–584. 37.Kleter, B., L. J. van Doorn, L. Schrauwen, A. Molijn, S. Sastrowijoto, J. ter
on May 16, 2020 by guest
http://jcm.asm.org/
Schegget, J. Lindeman, B. ter Harmsel, M. Burger, and W. Quint.1999. Development and clinical evaluation of a highly sensitive PCR-reverse hy-bridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol.37:2508–2517.
38.Kuipers, E. J., D. A. Israel, J. G. Kusters, M. M. Gerrits, J. Weel, A. van Der Ende, R. W. van Der Hulst, H. P. Wirth, J. Hook-Nikanne, S. A. Thompson, and M. J. Blaser.2000. Quasispecies development ofHelicobacter pylori
observed in paired isolates obtained years apart from the same host. J. Infect. Dis.181:273–282.
39.Kuipers, E. J., G. I. Perez-Perez, S. G. Meuwissen, and M. J. Blaser.1995.
Helicobacter pyloriand atrophic gastritis: importance of the cagAstatus. J. Natl. Cancer Inst.87:1777–1780.
40.Macarthur, M., G. L. Hold, and E. M. El-Omar.2004. Inflammation and cancer. II. Role of chronic inflammation and cytokine gene polymorphisms in the pathogenesis of gastrointestinal malignancy. Am. J. Physiol. Gastro-intest. Liver Physiol.286:G515–G520.
41.Marshall, D. G., A. Chua, P. W. Keeling, D. J. Sullivan, D. C. Coleman, and C. J. Smyth.1995. Molecular analysis ofHelicobacter pyloripopulations in antral biopsies from individual patients using randomly amplified polymor-phic DNA (RAPD) fingerprinting. FEMS Immunol. Med. Microbiol.10:
317–323.
42.Marshall, D. G., D. C. Coleman, D. J. Sullivan, H. Xia, C. A. O’Morain, and C. J. Smyth.1996. Genomic DNA fingerprinting of clinical isolates of Hel-icobacter pyloriusing short oligonucleotide probes containing repetitive se-quences. J. Appl. Bacteriol.81:509–517.
43.Martinez, A., C. Gonzalez, F. Kawaguchi, R. Montoya, A. Corvalan, J. Madariaga, J. Roa, A. Garcia, F. Salgado, H. Solar, and M. Palma.2001. [Helicobacter pylori: cagAanalysis andvacAgenotyping in Chile. Detection of a s2/m1 strain]. Rev. Med. Chil.129:1147–1153.
44.Mendez Castellano, H.1977. Proyecto Venezuela: estudio nacional de crec-imiento y desarrollo humanos de la Repu´blica de Venezuela. Ministerio de la Secretaria. FUNDACREDESA, Caracas, Venezuela.
45.Morales-Espinosa, R., G. Castillo-Rojas, G. Gonzalez-Valencia, S. Ponce de Leon, A. Cravioto, J. C. Atherton, and Y. Lopez-Vidal.1999. Colonization of Mexican patients by multipleHelicobacter pyloristrains with differentvacA
andcagAgenotypes. J. Clin. Microbiol.37:3001–3004.
46.Nomura, A., G. N. Stemmermann, P. H. Chyou, G. I. Perez-Perez, and M. J. Blaser.1994. Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann. Intern. Med.120:977–981.
47.Nomura, A. M., G. I. Perez-Perez, J. Lee, G. Stemmermann, and M. J. Blaser.2002. Relation betweenHelicobacter pylori cagAstatus and risk of peptic ulcer disease. Am. J. Epidemiol.155:1054–1059.
48.Odenbreit, S., J. Puls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas.
2000. Translocation ofHelicobacter pyloriCagA into gastric epithelial cells by type IV secretion. Science287:1497–1500.
49.Oona, M., M. Utt, I. Nilsson, O. Uibo, T. Vorobjova, and H. I. Maaroos.
2004.Helicobacter pyloriinfection in children in Estonia: decreasing sero-prevalence during the 11-year period of profound socioeconomic changes. Helicobacter9:233–241.
50.Peek, R. M., Jr., and M. J. Blaser.2002.Helicobacter pyloriand gastrointes-tinal tract adenocarcinomas. Nat. Rev. Cancer2:28–37.
51.Perez-Perez, G. I., D. Rothenbacher, and H. Brenner.2004. Epidemiology of
Helicobacter pyloriInfection. Helicobacter9(Suppl. 1):1–6.
52.Rodriguez-Larralde, A., J. Morales, and I. Barrai.2000. Surname frequency and the isonymy structure of Venezuela. Am. J. Hum. Biol.12:352–362. 53.Roosendaal, R., E. J. Kuipers, J. Buitenwerf, C. van Uffelen, S. G.
Meuwis-sen, G. J. van Kamp, and C. M. Vandenbroucke-Grauls.1997.Helicobacter pyloriand the birth cohort effect: evidence of a continuous decrease of infection rates in childhood. Am. J. Gastroenterol.92:1480–1482. 54.Scholte, G. H., L. J. van Doorn, A. Cats, E. Bloemena, J. Lindeman, W. G.
Quint, S. G. Meuwissen, and E. J. Kuipers.2002. Genotyping ofHelicobacter
pyloriin paraffin-embedded gastric biopsy specimens: relation to histological parameters and effects on therapy. Am. J. Gastroenterol.97:1687–1695. 55.Scholte, G. H., L. J. van Doorn, W. G. Quint, and J. Linderman.2001.
Genotyping ofHelicobacter pyloristrains in formalin-fixed or formaldehyde-sublimate-fixed paraffin-embedded gastric biopsy specimens. Diagn. Mol. Pathol.10:166–170.
56.Shortridge, V. D., G. G. Stone, R. K. Flamm, J. Beyer, J. Versalovic, D. W. Graham, and S. K. Tanaka.1997. Molecular typing ofHelicobacter pylori
isolates from a multicenter U.S. clinical trial byureCrestriction fragment length polymorphism. J. Clin. Microbiol.35:471–473.
57.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunst-mann, I. Dyrek, and M. Achtman.1998. Free recombination within Helico-bacter pylori. Proc. Natl. Acad. Sci. USA95:12619–12624.
58.Takata, T., R. Aras, D. Tavakoli, T. Ando, A. Z. Olivares, and M. J. Blaser.
2002. Phenotypic and genotypic variation in methylases involved in type II restriction-modification systems inHelicobacter pylori. Nucleic Acids Res.
30:2444–2452.
59.Taylor, N. S., J. G. Fox, N. S. Akopyants, D. E. Berg, N. Thompson, B. Shames, L. Yan, E. Fontham, F. Janney, F. M. Hunter, et al.1995. Long-term colonization with single and multiple strains of Helicobacter pylori
assessed by DNA fingerprinting. J. Clin. Microbiol.33:918–923.
60.Terry, K., S. Williams, and K. M. Ottemann.2004. Abstr. 104th Annu. Meet. Am. Soc. Microbiol., abstr. D-211, p. 229.
61.Tummuru, M. K., T. L. Cover, and M. J. Blaser.1993. Cloning and expres-sion of a high-molecular-mass major antigen ofHelicobacter pylori: evidence of linkage to cytotoxin production. Infect. Immun.61:1799–1809. 62.van Doorn, L. J., C. Figueiredo, F. Megraud, S. Pena, P. Midolo, D. M.
Queiroz, F. Carneiro, B. Vanderborght, M. D. Pegado, R. Sanna, W. De Boer, P. M. Schneeberger, P. Correa, E. K. Ng, J. Atherton, M. J. Blaser, and W. G. Quint.1999. Geographic distribution ofvacAallelic types of Helico-bacter pylori. Gastroenterology116:823–830.
63.van Doorn, L. J., C. Figueiredo, R. Rossau, G. Jannes, M. van Asbroek, J. C. Sousa, F. Carneiro, and W. G. Quint.1998. Typing ofHelicobacter pylori vacAgene and detection ofcagAgene by PCR and reverse hybridization. J. Clin. Microbiol.36:1271–1276.
64.van Doorn, L. J., C. Figueiredo, R. Sanna, S. Pena, P. Midolo, E. K. Ng, J. C. Atherton, M. J. Blaser, and W. G. Quint.1998. Expanding allelic diversity of
Helicobacter pylori vacA. J. Clin. Microbiol.36:2597–2603.
65.van Doorn, L. J., C. Figueiredo, R. Sanna, A. Plaisier, P. Schneeberger, W. de Boer, and W. Quint.1998. Clinical relevance of thecagA,vacA, andiceA
status ofHelicobacter pylori. Gastroenterology115:58–66.
66.van Doorn, L. J., Y. Henskens, N. Nouhan, A. Verschuuren, R. Vreede, P. Herbink, G. Ponjee, K. van Krimpen, R. Blankenburg, J. Scherpenisse, and W. Quint.2000. The efficacy of laboratory diagnosis ofHelicobacter pylori
infections in gastric biopsy specimens is related to bacterial density andvacA, cagA, andiceAgenotypes. J. Clin. Microbiol.38:13–17.
67.van Doorn, L. J., B. Kleter, L. Stuyver, G. Maertens, H. Brouwer, S. Schalm, R. Heijtink, and W. Quint.1994. Analysis of hepatitis C virus genotypes by a line probe assay and correlation with antibody profiles. J. Hepatol.21:122– 129.
68.Wirth, T., X. Wang, B. Linz, R. P. Novick, J. K. Lum, M. Blaser, G. Morelli, D. Falush, and M. Achtman.2004. Distinguishing human ethnic groups by means of sequences fromHelicobacter pylori: lessons from Ladakh. Proc. Natl. Acad. Sci. USA101:4746–4751.
69.Xu, Q., R. D. Morgan, R. J. Roberts, and M. J. Blaser.2000. Identification of type II restriction and modification systems inHelicobacter pylorireveals their substantial diversity among strains. Proc. Natl. Acad. Sci. USA97:
9671–9676.
70.Yamaoka, Y., E. Orito, M. Mizokami, O. Gutierrez, N. Saitou, T. Kodama, M. S. Osato, J. G. Kim, F. C. Ramirez, V. Mahachai, and D. Y. Graham.
2002.Helicobacter pyloriin North and South America before Columbus. FEBS Lett.517:180–184.