Acta Cryst.(2002). E58, o1177±o1179 DOI: 10.1107/S160053680201766X B. Sridharet al. C6H14N3O3+ClO4ÿ
o1177
organic papers
Acta Crystallographica Section E
Structure Reports Online
ISSN 1600-5368
L
-Citrullinium perchlorate
B. Sridhar,aN. Srinivasan,b
Bjoern Dalhuscand R. K.
Rajarama*
aDepartment of Physics, Madurai Kamaraj
University, Madurai 625 021, India,
bDepartment of Physics, Thiagarajar College,
Madurai 625 009, India, andcDepartment
of Chemistry, University of Oslo, Blindern, N-0315 Oslo, Norway
Correspondence e-mail: sshiya@yahoo.com
Key indicators
Single-crystal X-ray study
T= 105 K
Mean(C±C) = 0.001 AÊ
Rfactor = 0.023
wRfactor = 0.068
Data-to-parameter ratio = 32.1
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
#2002 International Union of Crystallography Printed in Great Britain ± all rights reserved
In the title compound, C6H14N3O3+ClO4ÿ, the citrullinium
residue forms a strong OÐH O hydrogen bonds with the
terminal O atom of a symmetry-related residue. This residue
has a gauche I-trans-trans-trans conformation. The crystal
structure is stabilized by an NÐH O hydrogen-bonding
network. The perchlorate anion is linked to the cation,
forming chains along theaaxis.
Comment
Citrulline amino acid is found in the urea cycle. The crystal
structures of l-citrulline hydrochloride (Naganathan &
Venkatesan, 1971), l-citrulline hydrochloride and l
-homo-citrulline hydrochloride (Ashidaet al., 1972), andl-citrulline (Toffoliet al., 1987) have been reported. In the present study,
the crystal structure determination of l-citrullinium
perchlorate, (I), was undertaken.
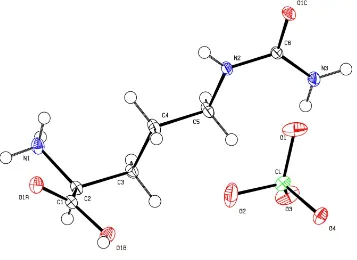
The asymmetric unit of the unit cell of (I) contains a citrullinium cation and a perchlorate anion (Fig. 1). The unsymmetrical CÐO bond distances [1.2189 (8) and 1.3151 (8) AÊ] and the OÐCÐC bond angles [122.81 (6) and
111.63 (5)] clearly con®rm the protonation of the carboxyl
group. Generally, the citrulline residue has three planar
groups,viz. the carboxyl group, the aliphatic group and the
carbamylamino group or urea unit (Naganathan &
Venka-tesan, 1971). The backbone conformation angle 1 (O1AÐ
C1ÐC2ÐN1) indicates a cis conformation [7.52 (8)]. The
deviation of the -amino N atom from the mean carboxyl
plane is 0.149 (1) AÊ. This tendency to twist about the CÐN
bond is found in various amino acids (Lakshminarayananet
al., 1967). The straight-chain conformation angle 1 (N1Ð
C2ÐC3ÐC4) is gauche I [68.99 (7)], while 2 (C2ÐC3Ð
C4ÐC5) istrans[ÿ177.39 (6)]. The other two conformation
angles3(C3ÐC4ÐC5ÐN2) and4(C4ÐC5ÐN2ÐC6) are
also bothtrans[ÿ179.47 (6) and 162.71 (7)]. The
conforma-tion angles51(C5ÐN2ÐC6ÐO1C) and52(C5ÐN2ÐC6Ð
N3) are 174.51 (6) andÿ4.72 (11), respectively. The aliphatic
chain has a fully extended planar conformation (Table 1). The average ClÐO bond distances and OÐClÐO bond
angles are 1.4450 (6) and 109.47 (4), respectively, con®rming
a nearly tetrahedral symmetry. The perchlorate anion plays a vital role in hydrogen bonding, stabilizing the crystal structure.
organic papers
o1178
B. Sridharet al. C6H14N3O3+ClO4ÿ Acta Cryst.(2002). E58, o1177±o1179The carboxyl O atom of the citrulline residue forms a strong
OÐH O hydrogen bond with the terminal O atom of a
symmetry-related residue (Table 2). The-,"- and-N atoms
(N1, N2 and N3) of the citrullinium residue form NÐH O
hydrogen bonds with the O atoms of the perchlorate anion. In
addition, the -N atom forms an intermolecular NÐH O
hydrogen bond with the terminal O atoms (Fig. 3). A class I hydrogen-bonding pattern is observed in the present structure,
having three two-center hydrogen bonds (Jeffrey & Saenger, 1991). Atom O4 of the perchlorate anion links the citrullinium
residues through NÐH O hydrogen bonds in a chain
running along the a axis [O4i H1AÐN1ÐH1B O4ii;
symmetry codes: (i)ÿx+ 2,y+1
2,ÿz+12; (ii)ÿx+ 1,y+ 1/2,
ÿz+1
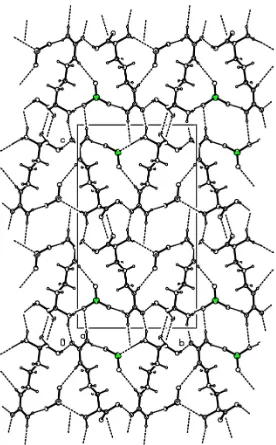
2]. The citrullinium residues are packed as corrugated
sheets in theabplane, interconnected by OÐH O hydrogen
bonding (Fig. 3).
Experimental
The title compound was crystallized by slow evaporation from an aqueous solution of citrulline and perchloric acid in a stoichiometric ratio of 1:1.
Crystal data
C6H14N3O3+ClO4ÿ
Mr= 275.65
Orthorhombic,P212121
a= 5.1113 (1) AÊ b= 11.3497 (2) AÊ c= 19.3853 (3) AÊ V= 1124.57 (3) AÊ3
Z= 4
Dx= 1.628 Mg mÿ3
Dm= 1.615 Mg mÿ3
Dmmeasured by ¯otation in a
mixture of carbon tetrachloride and xylene
MoKradiation Cell parameters from 7473
re¯ections
= 2.1±37.5 = 0.37 mmÿ1
T= 105 (2) K Block, colorless 0.700.450.30 mm
Data collection
Bruker SMART CCD diffractometer
!scans
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996)
Tmin= 0.77,Tmax= 0.89
25355 measured re¯ections
5866 independent re¯ections 5757 re¯ections withI> 2(I) Rint= 0.017
max= 37.5
h=ÿ8!8
k=ÿ19!18
l=ÿ32!33
Re®nement
Re®nement onF2
R[F2> 2(F2)] = 0.023
wR(F2) = 0.068
S= 1.03 5866 re¯ections 183 parameters
H atoms treated by a mixture of independent and constrained re®nement
w= 1/[2(F
o2) + (0.047P)2
+ 0.1185P]
whereP= (Fo2+ 2Fc2)/3
(/)max= 0.001
max= 0.44 e AÊÿ3
min=ÿ0.48 e AÊÿ3
Extinction correction:SHELXL97 Extinction coef®cient: 0.0217 (18) Absolute structure: Flack (1983) Flack parameter = 0.02 (3)
Figure 3
Packing diagram of (I), viewed down the c axis. H atoms have been omitted for clarity.
Figure 2
Packing diagram of (I), viewed down theaaxis.
Figure 1
Table 1
Selected geometric parameters (AÊ,).
O1AÐC1 1.2189 (8) O1BÐC1 1.3151 (8)
O1AÐC1ÐC2ÐN1 7.52 (8)
N1ÐC2ÐC3ÐC4 68.99 (7)
C2ÐC3ÐC4ÐC5 ÿ177.39 (6) C3ÐC4ÐC5ÐN2 ÿ179.47 (6)
C4ÐC5ÐN2ÐC6 162.71 (7)
C5ÐN2ÐC6ÐO1C 174.51 (6) C5ÐN2ÐC6ÐN3 ÿ4.72 (11)
Table 2
Hydrogen-bonding geometry (AÊ,).
DÐH A DÐH H A D A DÐH A
O1BÐH1 O1Ci 0.70 (2) 1.84 (2) 2.5292 (8) 171 (2)
N1ÐH1A O4ii 0.846 (17) 2.234 (17) 3.0226 (9) 155 (2)
N1ÐH1B O4iii 0.926 (15) 2.162 (16) 2.9699 (9) 145 (1)
N1ÐH1C O1Civ 0.918 (15) 1.906 (15) 2.7986 (8) 164 (1)
N2ÐH2A O2iii 0.837 (15) 2.583 (14) 3.3562 (10) 154 (1)
N3ÐH3C O4v 0.818 (13) 2.326 (13) 3.1205 (8) 164 (1)
N3ÐH3D O1vi 0.833 (15) 2.292 (15) 3.0952 (9) 162 (1)
Symmetry codes: (i) 1ÿx;yÿ1
2;12ÿz; (ii) 2ÿx;12y;21ÿz; (iii) 1ÿx;12y;12ÿz; (iv) 3
2ÿx;2ÿy;zÿ12; (v)12x;32ÿy;1ÿz; (vi) 1x;y;z.
All H atoms were located from a difference Fourier map. Those on the N and O atoms were re®ned freely, but the remainder were placed in idealized positions and were re®ned as riding on their parent atoms. 2484 Fridel pairs were measured and used.
Data collection:SMART(Bruker, 1998); cell re®nement:SAINT
(Bruker, 1998); data reduction: SAINT; program(s) used to solve
structure: SHELXS97 (Sheldrick, 1997); program(s) used to re®ne structure: SHELXL97 (Sheldrick, 1997); molecular graphics:
PLATON (Spek, 1999); software used to prepare material for publication:SHELXL97.
BS thanks the Council of Scienti®c & Industrial Research (CSIR), Government of India, for ®nancial assistance and RKR thanks the Department of Science and Technology (DST), Government of India, for ®nancial support. Financial support from UGC is acknowledged.
References
Ashida, T., Funakoshi, K., Tsukihara, T., Ueki, T. & Kakudo, M. (1972).Acta Cryst.B28, 1367±1374.
Bruker (1998).SMARTandSAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
Flack, H. D. (1983).Acta Cryst.A39, 876±881.
Jeffrey, G. A. & Saenger, W. (1991). Hydrogen Bonding in Biological
Structures. Berlin, Heidelberg, New York: Springer-Verlag.
Johnson, C. K. (1976).ORTEPII. Report ORNL-5138. Oak Ridge National Laboratory, Tennessee, USA.
Lakshminarayanan, A. V., Sashisekaran, V. & Ramachandran, G. N. (1967). In
Conformation of Biopolymers, edited by G. N. Ramachandran. London:
Academic Press.
Naganathan, P. S. & Venkatesan, K. (1971).Acta Cryst.B27, 1079±1085. Sheldrick, G. M. (1996).SADABS. University of GoÈttingen, Germany. Sheldrick, G. M. (1997). SHELXL97 and SHELXS97. University of
GoÈttingen, Germany.
Spek, A. L. (1999). PLATON for Windows. Utrecht University, The Netherlands.
Toffoli, P., Khodadad, P., Rodier, N. & Astoin, J. (1987).Acta Cryst.C43, 945± 947.
supporting information
sup-1
Acta Cryst. (2002). E58, o1177–o1179
supporting information
Acta Cryst. (2002). E58, o1177–o1179 [doi:10.1107/S160053680201766X]
L
-Citrullinium perchlorate
B. Sridhar, N. Srinivasan, Bjoern Dalhus and R. K. Rajaram
S1. Comment
Citrulline amino acid is found in the urea cycle. The crystal structures of L-citrulline hydrochloride (Naganathan &
Ventatesan, 1971), L-citrulline hydrochloride and L-homocitrulline hydrochloride (Ashida et al., 1972), and L-citrulline
(Toffoli et al., 1987) have been reported. In the present study, the crystal structure determination of L-citrullinium
perchlorate, (I), was undertaken.
The asymmetric part of the unit cell of (I) contains a citrullinium residue and a perchlorate anion (Fig. 1). The
unsymmetrical C—O bond distances [1.2189 (8) and 1.3151 (8) Å] and the O—C—C bond angles [122.81 (6) and
111.63 (5)°], clearly confirm the protonation of the carboxyl group. Generally, the citrulline residue has three planar
groups, viz. the carboxyl group, the aliphatic group and the carbamylamino group or urea fraction (Naganathan &
Venkatesan, 1971). The backbone conformation angle ψ1 (O1A—C1—C2—N1) is in a cis conformation [7.52 (8)°]. The
deviation of the α-amino N atom from the mean carboxyl plane is 0.149 (1) Å. This tendency of twisting about the C—N
bond is found in various amino acids (Lakshminarayanana et al., 1967). The straight-chain conformation angle χ1 (N1—
C2—C3—C4) is gauche I [68.99 (7)°], while χ2 (C2—C3—C4—C5) is trans [−177.39 (6)°]. The other two conformation
angles χ3 (C3—C4—C5—N2) and χ4 (C4—C5—N2—C6) are also both trans [−179.47 (6) and 162.71 (7)°]. The
conformation angles χ51 (C5—N2—C6—O1C) and χ52 (C5—N2—C6—N3) are 174.51 (6) and −4.72 (11)°, respectively.
The aliphatic chain has a fully extended planar conformation (Table 1).
The average Cl—O bond distances and O—Cl—O bond angles are 1.4450 (6) and 109.47 (4)°, respectively, confirming
a nearly tetrahedral symmetry. The perchlorate anion plays a vital role in hydrogen bonding and stabilize the crystal
structure. The carboxyl O atom of the citrulline residue forms a strong O—H···O hydrogen bond with its
symmetry-related terminal O atom (Table 2). The α-, ε- and η-N atoms (N1, N2 and N3) of the citrullinium residue form N—H···O
hydrogen bonds with the O atoms of the perchlorate anion. In addition, the α-N atom forms an intermolecular N—H···O
hydrogen bond with the terminal O atoms (Fig. 3). A class-I hydrogen-bonding pattern is observed in the present
structure, having three two-centered hydrogen bonding (Jeffrey & Saenger, 1991). Atom O4 of the perchlorate anion links
the citrullinium residues through Nα—H···O hydrogen bonds in a chain running along the a axis [O4i···H1A—N1—
H1B···O4ii; symmetry codes: (i) −x + 2, y + 1/2, −z + 1/2; (ii) (-x + 1, y + 1/2, −z + 1/2]. The citrullinium residues are
packed as corrugated sheets in the ab plane, interconnected by O—H···O hydrogen bonding (Fig. 3).
S2. Experimental
The title compound was crystallized by slow evaporation from an aqueous solution of citrulline and perchloric acid in a
supporting information
sup-2
Acta Cryst. (2002). E58, o1177–o1179
S3. Refinement
All H atoms were located from a difference Fourier map. Those on the N and O atoms were refined freely but the
remainder were placed in idealized positions and were refined as riding on their parent atoms. 2484 Fridel pairs were
[image:5.610.129.486.137.397.2]observed.
Figure 1
The molecular structure of the title compound, showing the atom-numbering scheme and 50% probability displacement
supporting information
sup-3
[image:6.610.168.444.67.512.2]Acta Cryst. (2002). E58, o1177–o1179
Figure 2
supporting information
sup-4
[image:7.610.127.484.71.265.2]Acta Cryst. (2002). E58, o1177–o1179
Figure 3
Packing diagran of (I), viewed down the c axis. H atoms have been omitted for clarity.
L-citrullinium perchlorate
Crystal data
C6H14N3O3+·ClO4−
Mr = 275.65
Orthorhombic, P212121
a = 5.1113 (1) Å b = 11.3497 (2) Å c = 19.3853 (3) Å V = 1124.57 (3) Å3
Z = 4 F(000) = 576
Dx = 1.628 Mg m−3
Dm = 1.615 Mg m−3
Dm measured by flotation in a mixture of carbon
tetrachloride and xylene Mo Kα radiation, λ = 0.71074 Å Cell parameters from 7473 reflections θ = 2.1–37.5°
µ = 0.37 mm−1
T = 105 K Block, colorless 0.70 × 0.45 × 0.30 mm
Data collection
Bruker SMART CCD diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
Detector resolution: 8.33 pixels mm-1
ω scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996) Tmin = 0.77, Tmax = 0.89
25355 measured reflections 5866 independent reflections 5757 reflections with I > 2σ(I) Rint = 0.017
θmax = 37.5°, θmin = 2.8°
h = −8→8 k = −19→18 l = −32→33
Refinement
Refinement on F2
Least-squares matrix: full R[F2 > 2σ(F2)] = 0.023
wR(F2) = 0.068
S = 1.03 5866 reflections 183 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
supporting information
sup-5
Acta Cryst. (2002). E58, o1177–o1179
w = 1/[σ2(F
o2) + (0.047P)2 + 0.1185P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max = 0.001
Δρmax = 0.44 e Å−3
Δρmin = −0.48 e Å−3
Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4
Extinction coefficient: 0.0217 (18)
Absolute structure: Flack H D (1983), Acta Cryst. A39, 876-881
Absolute structure parameter: 0.02 (3)
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based
on F, with F set to zero for negative F2. The threshold expression of
F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R
-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be
even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
supporting information
sup-6
Acta Cryst. (2002). E58, o1177–o1179
O1C 0.61697 (11) 1.04561 (5) 0.42560 (3) 0.01511 (9) N3 0.95763 (13) 0.91948 (5) 0.43683 (3) 0.01455 (9) H3C 0.928 (3) 0.9158 (11) 0.4782 (7) 0.015 (3)* H3D 1.061 (3) 0.8712 (12) 0.4194 (7) 0.017 (3)*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
Cl 0.01302 (6) 0.01252 (6) 0.01715 (6) −0.00012 (4) −0.00106 (5) 0.00302 (5) O1 0.0203 (3) 0.0156 (2) 0.0431 (4) 0.0045 (2) −0.0010 (3) −0.0050 (2) O2 0.0398 (4) 0.0334 (3) 0.0162 (2) −0.0069 (3) −0.0066 (2) 0.0089 (2) O3 0.0118 (2) 0.0219 (2) 0.0421 (3) 0.00055 (18) 0.0028 (2) 0.0052 (3) O4 0.0177 (2) 0.01562 (19) 0.0169 (2) −0.00234 (17) −0.00082 (17) 0.00517 (16) O1A 0.01091 (19) 0.0185 (2) 0.01366 (19) −0.00150 (15) −0.00222 (14) 0.00144 (16) O1B 0.0187 (2) 0.01492 (19) 0.0186 (2) −0.00439 (18) −0.00520 (16) 0.00358 (17) C1 0.0104 (2) 0.0129 (2) 0.00953 (19) −0.00050 (17) 0.00038 (16) −0.00213 (17) C2 0.0096 (2) 0.0141 (2) 0.00929 (19) −0.00007 (17) −0.00008 (16) −0.00218 (16) N1 0.0143 (2) 0.0173 (2) 0.01111 (19) −0.00463 (18) −0.00152 (17) 0.00068 (16) C3 0.0110 (2) 0.0154 (2) 0.0099 (2) 0.00152 (18) −0.00155 (16) −0.00241 (17) C4 0.0144 (3) 0.0229 (3) 0.0092 (2) 0.0058 (2) −0.00150 (18) −0.00383 (19) C5 0.0130 (3) 0.0203 (3) 0.0095 (2) 0.0038 (2) −0.00060 (17) −0.00397 (18) N2 0.0146 (2) 0.0148 (2) 0.00829 (18) 0.00438 (18) 0.00008 (16) −0.00087 (15) C6 0.0119 (2) 0.0115 (2) 0.0095 (2) 0.00113 (17) 0.00010 (16) −0.00131 (16) O1C 0.0169 (2) 0.0173 (2) 0.01119 (19) 0.00716 (16) 0.00089 (15) −0.00252 (15) N3 0.0167 (2) 0.0169 (2) 0.01003 (19) 0.00580 (19) −0.00088 (18) −0.00009 (16)
Geometric parameters (Å, º)
Cl—O1 1.4327 (7) C3—H3A 0.9700 Cl—O2 1.4360 (7) C3—H3B 0.9700 Cl—O3 1.4407 (6) C4—C5 1.5135 (9) Cl—O4 1.4706 (5) C4—H4A 0.9700 O1A—C1 1.2189 (8) C4—H4B 0.9700 O1B—C1 1.3151 (8) C5—N2 1.4578 (9) O1B—H1 0.70 (2) C5—H5A 0.9700 C1—C2 1.5164 (9) C5—H5B 0.9700 C2—N1 1.4946 (9) N2—C6 1.3443 (8) C2—C3 1.5308 (8) N2—H2A 0.837 (15) C2—H2 0.9800 C6—O1C 1.2670 (8) N1—H1A 0.846 (17) C6—N3 1.3436 (9) N1—H1B 0.926 (15) N3—H3C 0.818 (13) N1—H1C 0.918 (15) N3—H3D 0.833 (15) C3—C4 1.5279 (9)
supporting information
sup-7
Acta Cryst. (2002). E58, o1177–o1179
O2—Cl—O4 108.92 (4) C5—C4—H4A 109.2 O3—Cl—O4 107.82 (4) C3—C4—H4A 109.2 C1—O1B—H1 110.3 (17) C5—C4—H4B 109.2 O1A—C1—O1B 125.52 (6) C3—C4—H4B 109.2 O1A—C1—C2 122.81 (6) H4A—C4—H4B 107.9 O1B—C1—C2 111.63 (5) N2—C5—C4 109.21 (5) N1—C2—C1 108.25 (5) N2—C5—H5A 109.8 N1—C2—C3 111.02 (5) C4—C5—H5A 109.8 C1—C2—C3 114.19 (5) N2—C5—H5B 109.8 N1—C2—H2 107.7 C4—C5—H5B 109.8 C1—C2—H2 107.7 H5A—C5—H5B 108.3 C3—C2—H2 107.7 C6—N2—C5 125.34 (6) C2—N1—H1A 106.2 (12) C6—N2—H2A 116.0 (10) C2—N1—H1B 111.5 (9) C5—N2—H2A 118.4 (10) H1A—N1—H1B 110.4 (15) O1C—C6—N3 120.30 (6) C2—N1—H1C 111.4 (9) O1C—C6—N2 120.32 (6) H1A—N1—H1C 113.7 (15) N3—C6—N2 119.38 (6) H1B—N1—H1C 103.9 (13) C6—N3—H3C 117.6 (10) C4—C3—C2 113.42 (5) C6—N3—H3D 121.3 (9) C4—C3—H3A 108.9 H3C—N3—H3D 118.8 (13) C2—C3—H3A 108.9
O1A—C1—C2—N1 7.52 (8) C2—C3—C4—C5 −177.39 (6) O1B—C1—C2—N1 −174.58 (5) C3—C4—C5—N2 −179.47 (6) O1A—C1—C2—C3 131.73 (6) C4—C5—N2—C6 162.71 (7) O1B—C1—C2—C3 −50.37 (7) C5—N2—C6—O1C 174.51 (6) N1—C2—C3—C4 68.99 (7) C5—N2—C6—N3 −4.72 (11) C1—C2—C3—C4 −53.72 (8)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
O1B—H1···O1Ci 0.70 (2) 1.84 (2) 2.5292 (8) 171 (2)
N1—H1A···O4ii 0.846 (17) 2.234 (17) 3.0226 (9) 155 (2)
N1—H1B···O4iii 0.926 (15) 2.162 (16) 2.9699 (9) 145 (1)
N1—H1C···O1Civ 0.918 (15) 1.906 (15) 2.7986 (8) 164 (1)
N2—H2A···O2iii 0.837 (15) 2.583 (14) 3.3562 (10) 154 (1)
N3—H3C···O4v 0.818 (13) 2.326 (13) 3.1205 (8) 164 (1)
N3—H3D···O1vi 0.833 (15) 2.292 (15) 3.0952 (9) 162 (1)
Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) −x+2, y+1/2, −z+1/2; (iii) −x+1, y+1/2, −z+1/2; (iv) −x+3/2, −y+2, z−1/2; (v) x+1/2, −y+3/2, −z+1; (vi) x+1,