Structure, Expression
and
Phylogenetic
Analysis of
the Gene Encoding
Actin
I
in
Pneumocystis
carinii
Leah D. Fletcher,*
JohnM. McDowell,t Richard R. Tidwell,” Richard B. Meaghert and
Christine C. Dykstra”
*Department of Pathology, School of Medicine, University of North Carolina, Chapel Hill, North Carolina 27599, and +Department of Genetics, University of Georgia, Athens, Georgia, 30602
Manuscript received September 27, 1993 Accepted for publication March 23, 1994
ABSTRACT
Actin is a major component of the cytoskeleton and one of the most abundant proteins found in eukaryotic cells. Comparative sequence analysis shows that this essential gene has been highly conserved throughout eukaryotic evolution making it useful for phylogenetic analysis. Complete cDNA clones for the actinencoding gene were isolated and characterized from Pneumocystis carinii purified from immunosuppressed rat lungs. The nucleotide sequence encodes a protein of 376 amino acids. The pre- dicted actin protein of P. carinii shares a high degree of conservation to other known actins. Only one major actin gene was found in P. carinii. The P. carinii actin sequence was compared with 30 other actin sequences. Gene phylogenies constructed using both neighbor-joining and protein parsimony methods places the P . carinii actin sequence closest to the majority of the fungi. Since the phylogenetic relationship
of P. carinii to fungi and protists has been questioned, these data on the actin gene phylogeny support
the grouping of P. carinii with the fungi.
A
CTIN genes are descended by duplication and di- vergence from common ancestral genes and arose early in eukaryotic evolution. Crucial cellular processes influenced by actin include motility, regulation of cell growth and differentiation, endocytosis, exocytosis and structural stability (POLLARD 1990; POLLARD and COOPER 1986; STOSSEL 1984). Approximately six different iso- forms of actin are expressed in warm-blooded verte- brates and plants. Many of these actin isoforms appear to be developmentally regulated and also exhibit a cell type-specific expression (MCLEAN et al. 1990). Recent studies have shown that several lower eukaryotes also contain multiple actin isoforms or actin-related pro- teins ( i . e . , Plasmodium falciparum (WESSELING et al. 1988a,b; 1988), Schizosaccharomycespombe (LEES-MILLER et al. 1992), and Saccharomyces cerevisiae (SCHWOB and MARTIN 1992) ).
P. carinii has been a challenge to taxonomists for more than
70
years. It was originally classified as a pro- tozoan based on its misidentification as a trypanosome (CHAGAS 1909) and its susceptibility to anti-protozoal agents ( i. e., pentamidine, berenil and primaquine) and resistance to anti-fungal agents. Although some life cycle stages of P. carinii are similar in appearance to those ofprotozoans like Toxoplasma gondii, recent studies at the molecular level have suggested that P. carinii may be more closely related to fungi in the Ascomycota group (EDMAN et al. 1988). Others have suggested a closer association with the ‘‘F&izopoda/Myxomycota/ Zygomycota” group ( W A T A N ~ E et al. 1989) than with protozoa. Studies comparing the 18s rRNA sequence
Genetics 157: 743-750 uuly, 1994)
from P . cariniiwith eight other taxa showed the greatest amount of homology 18s rRNA sequences from fungi such as S. cerevisiae and Neurospora crassa (CUSHION et al. 1988). Additional evidence for a fungal classifica- tion is demonstrated by the fact that the dihydrofolate reductase and thymidylate synthase genes of P. carinii are separate and not encoded by a bifunctional enzyme
0.
C. EDMAN et al. 1989; U. EDMAN et a l. 1989). Generally, the fungi, unlike protozoans, have the two enzyme ac- tivities on separate polypeptides. Comparisons of gene sequences could help resolve the debate over the clas- sification of P. carinii. The ability to classify this organ- ism has significant clinical relevance for the determina- tion of new therapeutic agents.We have completely sequenced cDNA clones contain- ing the major actin gene. Since the amino acid sequence and biochemical properties of this protein have been highly conserved throughout evolution, actin is an ideal gene for phylogenetic analysis. P. carinii appears to have only one major actin gene. We have also identified a second actin-related gene with homology to mamma- lian centractin (L. D. FLETCHER and C. C. DYKSTRA, manu- script in preparation). This study describes the isolation and characterization of the P. carinii actin I gene and the phylogenetic analysis of the nucleotide and derived amino acid sequences.
MATERTALS AND METHODS
744 Leah D. Fletcher et al.
trophozoites by methods previously described (FLETCHER et al.
1993).
Oligonucleotide primers: The primer sequences were based on known conserved regions of actin proteins from sev- eral organisms and designed with the P. carinii A
+
T codon bias described by FLETCHER et al. (1993). The actin primer pairs were 5'dGG GAT GAT ATG GAA AAA AT(T/A) TGG C and(A/T)AC, corresponding to nucleotides 239-262 and 490-512, respectively, of the P. falciparum actin I gene (WESSELING et al. 1988a). Oligonucleotides were synthesized by the University of North Carolina, Department of Pathology core facility.
Polymerase chain reaction and cloning of amplified prod- ucts: The polymerase chain reactions (PCR) were performed in a BioTherm thermal cycling oven. Thirty rounds of ampli- fication were performed by denaturation at 92", annealing at 48", and elongation at 72" (the duration of each step was 1 min). Each amplification reaction contained 100 ng of genomic P. carinii DNA. Rat testes and S . cerevisiae DNA samples were used as negative controls. P. carinii PCR prod- ucts were isolated from a 1% low melting point agarose gel by GlasPak (National Scientific Supply Company) procedures. Purified PCR products were ligated directly into a pBluescript I1
K
S
'
(Stratagene) T-tailed vector according to MARCHUK et al.(1991). Ligated DNA was transformed into Escherichia coli
(DH5-a) cells by electroporation at 25 pF, 200 ohms, and 25 kV (Bio-Rad Cell-Porator)
.
Bacteriawere allowed to recover for 1 hr at 37" with shaking, before selection on LB plates con- taining ampicillin (50 pg/ml), with X-gal+
isopropyl PO- thiogalactopyranoside, permitting blue us. white recombinant selection. Plasmid DNA was isolated from the recombinants by the alkaline lysis method (SAMBROOK et al. 1989). Recombinant DNA fragments were digested from the vector with PstI/Hind111 restriction endonucleases and isolated from 1% low melting point agarose gels by the GlasPak (National Scientific Supply Company) protocol. The recovered DNA was used for random primed DNA labeling (Boehringer Mannheim) with
[32P]dCTP (Amersham).
Isolation of the actin I cDNAs: A hgtll P. carinii cDNA library (kindly provided by J. A. FISHMAN, Infectious Disease Unit, Massachusetts General Hospital, 149 13th Street, Charlestown, Massachusetts 02129), was screened with the 32P- labeled actin gene fragment described above. Three clones, (41-1, 6-4-1 and El-1) were selected and purified by two
rounds of single plaque isolation and rescreening. The EcoRI
fragments from each of the cDNA clones was isolated, sub- cloned into the pBluescript I1
K
S
'
(Stratagene) vector and sequenced.Nucleic acid hybridization: Restriction endonucleases were used according to the manufacturers' instructions. Genomic
P. carinii, S. cerevisiae and rat testes DNAs were digested with a variety of endonucleases, fractionated by agarose gel elec- trophoresis and blotted onto nitrocellulose membranes ac- cording to SOUTHERN (1975). Hybridizations were performed overnight at 55-60" in 6 X SSC, 1 X Denhardt's solution and 50 pg/ml salmon sperm DNA, after adding the '*P-labeled actin probe. Following hybridization, filters were washed once in 2 X SSC/O.l% sodium dodecyl sulfate (SDS) for 5 min at room temperature and three timesin 0.1 X SSC/O.l% SDS for 10 min each at 55-60". Filters were exposed to Kodak XARfilm (Rochester, New York).
Isolation and purification of P. carinii mRNk P. c a r i n i i mRNA was isolated using a FastTrack mRNA isolation kit (In- vitrogen Corp., San Diego, California). Two different sources of organisms were utilized: (1) cysts and trophozoites purified as described by FLETCHER et al. (1993) and frozen in l-g quan- 5'd-GC ATA (A/T/C)CC CTC ATA (A/T)AT (A/T)GG
TABLE I
Actin sequences used for phylogenetic analysis
Name
GenBank Abbreviation accession no.
Pneumocystis carinii
Absidia glauca Aspergillus nidulans Candida albicans Kluyveromyces lactis Saccharomyces bayanus Saccharomyces cerevisiae Schizosaccharomyces pombe Phytophthora infestans Phytophthora megasperma
Thermomyces lanuginosus
Acanthamoeba castellani Achlya bisexualis Cryptosporidium paruum Dictyostelium discoideum Entamoeba histolytica Naegleria fowleri Oxytrichia nova Physarum polycephalum Plasmodium falciparum Tetrahymena thermophila Trypanosoma brucei
Volvox carteria
Oryza sativa
Piastrer ochraceus Drosophila melanogaster
Caenorhabditis elegans
Gallus gallus pactin Rattus rattus a-actin Homo sabiens a-actin Fungi: Protists: Green algae: Plant: Invertebrate: Vertebrate:
Pea L2183
A d M64729
A n i M22869
Cal X16377
Kla M25826
Sba Dl2534
See LO0026
P i n S37672
Pme S5 1076
T l a X07463
Aca v00002
Abi X59936
Ddi X03284
Ehi MI9871
Ono M22480
S P O YO0447
CPa M86241
Nfo M90311
4 0 M21501
Pfa M19146
T t h M 13939
Tbr M20310
Vca M33963
Osa X 16280
POC M26501
Dme KO0670
Cae X1 6796
Gga LO8 165
Rra X06801
Hsa 105192
~~
tities at -80" until needed and (2) P. carinii-infected rat lung tissue excised, immediately frozen in liquid nitrogen (1-g quantities) and stored at -80" until needed. P. carinii mRNA was isolated from these two different sources using the Fast- Track protocol. Contaminating DNA was removed with RQ1 RNase-free DNase I (Promega). The mRNA samples were stored in elution buffer at -20" after the addition of 20 units RNasin (Promega).
P. carinii Actin I 745
23.1
-
9.4-
6.6-
4.4-
2.3
-
2.0-
FIGURE 1 . 4 o u t h e r n blot analysis of the P. carinii actin gene. The actin I-specific DNA fragment was isolated from from clone 41-1 and used for random primed DNA labeling
(Boehringer Mannheim) with [32P]dCTP (Amersham) and hy- bridized to the filter as described in the MATERIAIS AND METHODS. Lanes 1 and 4 contain genomic P. carinii DNA digested with HincII and EcoRV, respectively; lanes 2 and 5 contain genomic S. cerevisiae DNA restriction digested with H i n d and EcoRV, respectively; and lanes 3 and 6 contain genomic rat testes DNA restriction digested with HincII and EcoRV, respectively.
twice. Filters were covered with Saran-Wrap and exposed to Kodak X A R film.
DNA sequencing: The DNA sequence of the actin gene was generated with a Sequenase kit (U.S. Biochemical Corp.) and [a-%]dATP (Amersham) on selected subclones. The se- quences were aligned and analyzed with the Macvector (IBI)
and GCG (DEVEREUX et al. 1984) programs.
Species and GenBank accession numbers of actin genes used used for phylogenetic analysis: Table 1 lists the 31 spe- cies, abbreviations, and GenBank accession numbers used in the actin phylogenetic analysis. Examples from six kingdoms were chosen. Every protozoan and most of the fungal actin sequences available in GenBank were used for the analysis.
RESULTS AND DISCUSSION
Identification, isolation, and nucleotide sequence of P. carinii actin genes: Actin is a ubiquitous single polypeptide chain of about 375 residues. The amino acid sequence and biochemical properties of actin have been highly conserved throughout evolution (VANDEKERCKHOVE
and WEBER 1984). Two criteria were employed to selec- tively identify the P. carinii actin gene(s). Previous stud- ies have suggested that P. carinii has a strong preferred codon usage that could be employed for P . carinii gene identification (EDMAN et al. 1989; FLETCHER et al. 1993). Thus, we designed A
+
T-biased degenerate oligonucle- otide primers to specifically amplify the P. carinii actin gene by PCR techniques. Secondly, the regions of the actin consensus sequence selected for primer design in- corporated the amino acids that have shown the least1 2 3
kb
2.4 -.
1.3 "
FIGURE 2.-Northem analysis of P. carinii mRNA. Poly(A+) RNA isolated from either purified cysts and trophozoites (lane 1) or infected rat lung tissue (lanes 2 and 3) was electrophe resed and blotted onto nitrocellulose as described in MATERIALS AND METHODS. The filters were hybridized with DNA from actin
I clone (41-1). RNA size marker positions are indicated on the left.
degeneracy in their codon selection as much as possible (FLETCHER et al. 1993). This approach optimizes primer hybridization to
P.
carinii actin sequences and reduces the chances of hybridization to other actin-related genes. The actin primers amplified two fragments from genomicP.
carinii DNA, one of 263 bp and the other -350 bp (data not shown). Amplification of genomicS.
cerevisiae DNA and rat testes DNA with the same prim-
ers, yielded no amplified DNA, confirming the utility of this approach. DNA sequence analysis of recombinants containing the PCR-amplified products revealed that the 263-bp fragment (actin I) had significant homology to vertebrate cytoplasmic
( p )
actin while the -350-bp fragment (actin 11) had less than 60% homology to ver- tebrate actin, but appeared to be an actin-relatedsequence (L. D. FLETCHER, L. CHRISTOPHER and C. C. D w m , in preparation). The actin
I
PCRgenerated fragment was radiolabeled and used to screen a hgtll cDNA library. DNA sequence analysis and restriction mapping of subclones for three individual Agtl 1 isolates (41-1, 6-41, and El-1) indicated that they were iden- tical to the actin I PCRfragment. ActinI
clones 41-1 and 64-1 were full length cDNA clones and R-1-1 was a shorter (-two-thirds the size) cDNA clone that mapped to the 3' end of the gene. The completeP.
carinii actin I DNA sequence was obtained from these three clones746 Leah D. Fletcher et al.
Pileup start
:1
1 MEDEIAALVIDNGSGMCKFAGDDAPRAVFPSIVGRPREIQGIMVGMGQKDSYVGDEAQS MDDD---V---_---V--- MEEE---H---_-- MEEEV---H---I--- MDSEV---
MGEEWQ---V---NV---V---S---KNP---EE--AF---T
DGEDVQ---_---T-V---
61 KRGILTLKYPIEHGIVSNW~DMEKIWHHTFYNELRVAPE~HPALLTEAPLNPKSNREKMT ---T---V---A--- ---N---C---
---R---V-T---V---I---
---R---T---V---M--- ---T---V---A--- ---.---T---_---A---V---G---R--
121 QIMFETFNTPAFWAIQAV~SLYASGRTTGIVLDSGDG~HTVPIYEGYALPHAILRLNL ---M---M---D- --I---A---M--D-
- -V- - - _ -V-- "-S
_ _
- _ - - - _ _ -_ _
- _ - - - - - - - - - "-V-_ _ _ _
-F- - - - - -S -VDM FrGURE 3"comparison Of p' carinii actin I-
_ _ _ _ _ _
-v_ _ _ _
-s--_ _ _
- - -s- - - -_ _ _ _
-_ _ _
- - - --v- - --A-FS-- - - - _ _ ID- with actins from other species. The amino acid-
_ _ _ _
-_ _
- _ "M-_ _ _ _
-_ _ _
-_ _ _ _ _
-_ _
"M_ _ _ _ _ _
s - - - -- _ _ _ _ _
-_ _ _
- - - -D- sequence of P. carinii actin I was aligned with181
AGRDLTDYLMKILTERGYNTTTAEREIVRDIKERLCYVLDFEQEIHTASSSSSLEKSY
and tin (KOST GALLwTZ et al. 1987), 1983), A . S. nidulans actin pombe actin (MERTINS (FIDEL_ - - - _ - _ _ M
_ _ _ _
TFS_ _ _ _ _ _ _ _ _ _ _ _ _
K- - - _ _ _ _ _ _ _ _ LQ--AQ_ _ _ _ _ _ _ _
et al. 1988), S. cerevisiae actin ( N G and_ _ _ _ _ _ _ _ _ _ _ _ _ A
_ _ _ _
TFS_ _ _ _ _ _ _ _ _ _ _ _ _
K - - _ _ _ _ _ _ _ _ - _ o"-o_ _ _ _ _ _ _ _
ABELSON 1980), D. discoideum actin (ROWS---S--V--M---S---S---M--D- the sequences of G . gallus cytoplasmic (p) ac-
---_---SF---_---"---"-"---
..
---s----sFS----__---K-"K---MQ--AQ-_-I----
---M---SF---K-A---~--A---A--- ---E---H----GFS-S--K---K---I--N-DE-MK-SEQ--DI----
.~ - "
and FIRTEL 1985), and P . falciparum actin I
(WESSELING et al. 1988a). Residues which are identical to the P. carinii protein are indicated 2 4 1 ELPDGQVITIGNERFRAPEALFQPSIVGMETCGIHETTF~SIMKCDVDIRKDLYSNIVMS by dashes (-)7 while differences are shown by
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
c_ _ _ _ _ _ _ -
FL"-S- _ _ _ - _ _ _ _ _ _ _ _ _ - _ _ _ _ _ _ _ _
A-T-L- the amino acid which occurs in the other ---AL-L-NA----A-~---G-~--- proteins.---K---VL-L-SG---V---I---V---G---
---H--vL"-sA"DQ--DQ--y-_---V--E--G---
---C---FL---SA---Y---G-V-L-
---NI--V---C---FL-K-AA---T---K---G---L-
301 GGTTMYPGIADRMQKEITA~APSSMKIKIVAPPERKYS~IGGSILASLSTFQQ~ISKQ
---_---T---I---
---Q---V--- -""-"-s""""""""v"I"""""""""""""""
---F----E---V--I---T--- ---F---N--L---T---I---_---E ---E-TGE-LTRD--T----T----V---S---T-E
361 EYDENGPSIVYRKCF* P. carinii "--s""-H""- G. gallus (p)
"--S--G"""-- S. pombe -"-s""-H""- A. nidulans
-"-S-""HH"" S. cerevisiae
""S""-H""- P. falciparum
-"-s""-H""- D. discoideum
striction mapping and sequencing strategy will be pro- rinii actin gene. Since any purification of P. carinii has
vided upon request. the potential to remove yet unidentified forms that may
To confirm that the recombinants were derived from contain life cycle stage-specific actin-like mRNAs, we
P. carinii DNA, the actin I gene coding region was hy- isolated mRNA directly from whole, quick frozen, bridized to a Southern blot of restriction digested P. carinii-infected rat lungs as well as from purified genomic P. carinii,
S.
cerevisiae and rat testes DNA (Fig-ure 1). Specific hybridization to genomic P. carinii DNA was observed for the actin I gene. The second, fainter band in the hybridization to the EcoRV digest was o b served due to the presence of an EcoRV site in an intron at the 5' end of the genomic DNA. No specific hybrid- ization was detected in the yeast or rat DNA controls. The hybridization results confirm that the clones origi- nated from P. carinii DNA.
Detection of actin I mRNk Northern blotting was performed to determine the transcript size of the P. ca-
cysts and trophozoites.
P . carinii Actin I
on0 Tbr
\
\
Ddi
747
113
A.
Osa
58
32
Tbr
211 Trh
I I3 25 Osa
32 Nfo
-
26 pfaCPa
l 3
Abi
13 34
,
810 Pi"Hsa
Pme
I I Ca;&
" Dme 3l
16 Ehi
4 0
19 l o Aco
7 680
I 1 E H a l
-
POCRra 23
19 Pca
22 spO
15
23 T , ~
Ani
FIGURE 4.-Gene trees for actin se- quences. The branches are drawn to scale with the distance values placed in each node for both trees. In ad- dition, for both trees the protists are generally grouped at the top and the fungi are grouped towards the bot- tom. (A) A gene tree relating 28 ac- tin sequences from six eukaryotic kingdoms was constructed by the neighborjoining method (SAITOU and NEI 1987). The neighbor- joining distances correlate approxi- mately with the fraction of RNS between sequences. This is an un- rooted tree. The members of the Ascomycetes have been underlined.
(B) A consensus gene tree relating
30 actin protein sequences from six kingdoms to that of P . carinii was constructed by the PAUP method (SWOFFORD 1991) utilizing the heu- ristic and MULPARS options with
bootstrapping.
B.
with its high sensitivity, did not amplify any bands (data not shown).
Predicted amino acid sequence comparisons: A comparison of the deduced amino acid sequence with the GenBank sequence data revealed several actin proteins with high conservation to the P. carinii actin I including; Rattus rattus p-actin (93% identity), S. pombe (91% identity), Aspergillus nidulans (95%
identity), S. cermisiae (94.7% identity), Dictyostelium discoideum (89% identity) and Plasmodium falciparum
actin
I
(88.8% identity). These sequences are shown in Figure 3, lined up to illustrate their sequence conservation.Phylogenetic analysis of P. carinii actin I: Since the taxonomic assignment of P. carinii has been contro- versial and actin is commonly used for phylogenetic analysis, a major goal of this study was to determine the evolutionary relationship of P. carinii actin I to actins
from other eukaryotes with particular attention devoted to members of the fungi
and
protozoa. Thirty se- quences, listed in Table 1, from organisms representing six eukaryotic kingdoms were included in the analysis with P. carinii. Only the coding sequence of each or- ganism was used. To optimize the alignment, each se- quence alignment began with the codon homologous to alanine 7 of P. carinii actin I and a one-codon gap was introduced at nucleotides 812-814 to accommodate theextra codon found in Trypanosoma brucei and
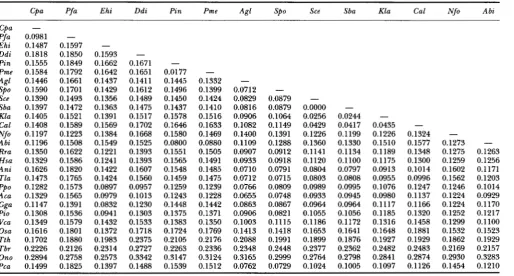
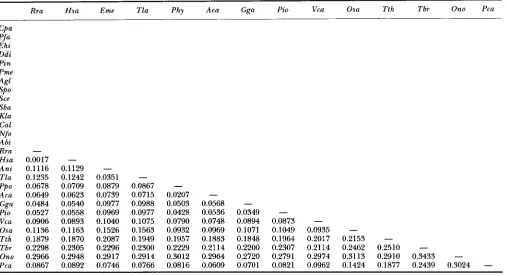
Oxytrichia nova. A distance matrix of corrected replace- ment nucleotide substitution (RNS) rates was calculated according to the method of LI et al. (1985) (Table
2)
for 28 of the sequences. RNS is a measure of the nucleotide changes that result in amino acid changes.748 Leah D. Fletcher et al.
TABLE 11
Matrix of replacement nucleotide substitution data between pairs of actin genes
Cpa Pfa Ehi Ddi Pin Pme Agl Spo Sce Sba Kla Cal Nfo Abi
Cpa -
Pfa 0.0981 -
Ehi 0.1487 0.1597 -
Ddi 0.1818 0.1850 0.1593 -
Pin 0.1555 0.1849 0.1662 0.1671 -
Pme 0.1584 0.1792 0.1642 0.1651 0.0177 -
Agl 0.1446 0.1661 0.1437 0.1411 0.1445 0.1332
-
Sce 0.1390 0.1493 0.1356 0.1489 0.1450 0.1424 0.0829 0.0879
-
Sba 0.1397 0.1472 0.1363 0.1475 0.1437 0.1410 0.0816 0.0879 0.0000 -
Kla 0.1405 0.1521 0.1391 0.1517 0.1578 0.1516 0.0906 0.1064 0.0256 0.0244 -
Cal 0.1408 0.1589 0.1569 0.1702 0.1646 0.1633 0.1082 0.1149 0.0429 0.0417 0.0435 -
Abi 0.1196 0.1508 0.1549 0.1525 0.0800 0.0880 0.1109 0.1288 0.1360 0.1330 0.1510 0.1577 0.1273
-
Rra 0.1350 0.1622 0.1221 0.1393 0.1551 0.1505 0.0907 0.0912 0.1141 0.1134 0.1189 0.1348 0.1275 0.1263
Hsa 0.1329 0.1586 0.1241 0.1393 0.1565 0.1491 0.0933 0.0918 0.1120 0.1100 0.1175 0.1300 0.1259 0.1256
Ani 0.1626 0.1820 0.1422 0.1607 0.1548 0.1485 0.0710 0.0791 0.0804 0.0797 0.0913 0.1014 0.1602 0.1171
Tla 0.1473 0.1765 0.1424 0.1560 0.1459 0.1475 0.0712 0.0715 0.0803 0.0808 0.0955 0.0996 0.1562 0.1203
Ppo 0.1282 0.1573 0.0897 0.0957 0.1259 0.1239 0.0766 0.0809 0.0989 0.0995 0.1076 0.1247 0.1246 0.1014
Aca 0.1329 0.1565 0.0979 0.1013 0.1243 0.1228 0.0655 0.0748 0.0933 0.0945 0.0980 0.1137 0.1224 0.0929
Gga 0.1147 0.1391 0.0832 0.1230 0.1448 0.1442 0.0863 0.0867 0.0964 0.0964 0.1117 0.1166 0.1224 0.1170
Pi0 0.1308 0.1536 0.0941 0.1303 0.1375 0.1371 0.0906 0.0821 0.1055 0.1056 0.1185 0.1320 0.1252 0.1217
Vca 0.1349 0.1579 0.1432 0.1533 0.1383 0.1350 0.1003 0.1115 0.1186 0.1172 0.1316 0.1458 0.1299 0.1100
Osa 0.1616 0.1801 0.1372 0.1718 0.1724 0.1769 0.1413 0.1418 0.1653 0.1641 0.1648 0.1881 0.1532 0.1523
Tth 0.1702 0.1880 0.1983 0.2375 0.2105 0.2176 0.2088 0.1991 0.1899 0.1876 0.1927 0.1929 0.1862 0.1929
Tbr 0.2226 0.2126 0.2314 0.2727 0.2263 0.2336 0.2348 0.2448 0.2377 0.2362 0.2482 0.2483 0.2169 0.2157
Ono 0.2894 0.2758 0.2573 0.3342 0.3147 0.3124 0.3165 0.2999 0.2764 0.2798 0.2841 0.2874 0.2930 0.3283
Pca 0.1499 0.1825 0.1397 0.1488 0.1539 0.1512 0.0762 0.0729 0.1024 0.1005 0.1097 0.1126 0.1454 0.1210
SPO 0.1590 0.1701 0.1429 0.1612 0.1496 0.1399 0.0712
-
Nfo 0.1197 0.1223 0.1384 0.1668 0.1580 0.1469 0.1400 0.1391 0.1226 0.1199 0.1226 0.1324
-
of taxa and iteratively clusters them into nodes which minimizes the total branch length, producing a “mini- mum evolution” tree. This tree is shown in Figure 4 A .
The fungal, protist, green algal, plant and animal actin sequences generally form distinct groups in the tree. The P. carinii actin is most closely related to most of the fungal actins, but away from most of the ascomycetous actins. To confirm these results, the amino acid se- quences of each of the
28
organisms were aligned by the GCG Pileup program (DEVEREUX et al. 1984) and used in a “Protpars” (Phylip 2.9) protein parsimony program. Twelve trees were produced (data not shown), all of which were very similar in topology to the neighbor- joining tree. The only difference observed from the twomethods was that in 3 of the 12 trees, the order of S . pombe and Absidia glauca were switched. Each of the 12 trees indicated that P. carinii actin was most closely related to the fungal actin sequences.
As
a further test, the protein parsimony method described by SWOFFORD (1991) was utilized to perform a second protein parsi- mony analysis on the original 28 sequences with two ad- ditional sequences added (Drosophila melanogaster andCaenorhabditis elegans)
.
This analysis was performedheuristically with the bootstrapping option The results are shown in Figure 4B as a cladistic tree. Thus three different methods of phylogenetic analysis placed the
P. carinii actin with the fungal actins. While it has been suggested that P. carinii should be placed among the ascomycetous fungi, S. pombe is the only ascomycete ac- tin to be closely related to the P. carinii actin. The next
closest fungal actin to that from P. carinii is the actin of A. nidulans.
It is also interesting to note that two oomycetous spe- cies actins (Phytophthora megasperma and Phytophthora
infestins) and one zygomycete actin (Achlya bisexualis)
were found in the middle of the protist region of both the neighbor-joining and protein parsimony trees. These three species are poorly described fungi, so more information is required to determine whether this is a case for convergent evolution or misplacement of these organisms with the fungi.
CONCLUSION
Designing probes or primers based on known con- served regions of actin proteins from other organisms, coupled with codon preference information facilitated the isolation of the P. carinii actin I gene. Complete cDNA clones for the actin I gene have been se- quenced. The P. carinii actin I is highly conserved with other known actins and phylogenetic analysis of this gene by two independent methods places the P . carinii actin with fungal actins, closest to that of
S.
P. carinii Actin I
Rra Hsa Eme Tla Phy Aca Gga Pi0 Vca Osa Tth Tbr Ono Pca
CPa Pfa Ehi Ddi Pin Pme 4 1
SPO
Sce Sba Kla
Gal
Abi
Rra -
Hsa 0.0017 -
A n i 0.1116 0.1129 -
Tla 0.1235 0.1242 0.0351 -
Aca 0.0649 0.0623 0.0739 0.0715 0.0207 -
Gga 0.0484 0.0540 0.0977 0.0988 0.0503 0.0568 -
Vca 0.0906 0.0893 0.1040 0.1075 0.0790 0.0748 0.0894 0.0873 -
Osa 0.1136 0.1163 0.1526 0.1563 0.0932 0.0969 0.1071 0.1049 0.0935
-
Tbr 0.2298 0.2305 0.2296 0.2300 0.2229 0.2114 0.2200 0.2307 0.2114 0.2462 0.2510 -
Ono 0.2966 0.2948 0.2917 0.2914 0.3012 0.2964 0.2720 0.2791 0.2974 0.3113 0.2910 0.3433 -
Pca 0.0867 0.0892 0.0746 0.0766 0.0816 0.0609 0.0701 0.0821 0.0962 0.1424 0.1877 0.2439 0.3024
-
Nfo
PPo 0.0678 0.0709 0.0879 0.0867
-
Pi0 0.0527 0.0558 0.0969 0.0977 0.0428 0.0536 0.0349 -
Tth 0.1879 0.1870 0.2087 0.1949 0.1957 0.1883 0.1848 0.1964 0.2017 0.2153 -
TAYLOR and BOWMAN (1993) suggest that the closest known relatives of P. carinii are most likely the ascomy- cete fungi rather than the basidiomycetous red yeasts as
suggested by WAKEHELD et aL (1992).
An
analysis of many more actins (and similar studies with other genes) is required to fully resolve this ques- tion. For example, P. carinii could easily be classified in the Mycetozoan class based on cytological features of encysted cells or spores (S. H. HUTNER in LEE et al. 1985), but among the Mycetozoans, only actins from the orders Dictyosteliida and Physarida within this class have been cloned. P. carinii shares some physical characteristics with the cyst and trophic stages of some members of the orders Protosteliida, the Guttulinala and the Plasmodio- phoridae. The taxonomy of the Mycetozoans is even more controversial as the protostelids, plasmodiopho- rids, cellular slime molds and myxomycetes have been grouped together merely for convenience ( O L ~ 1975; LEVINE et al. 1980). Their unifying characteristics are the presence of stalked fruiting bodies (with the ex- ception of the plasmodiophorids) and plasmodial veg- etative stages in all groups except the cellular slime molds, which form pseudoplasmodia. While P. cam’niihas not been described as containing a “plasmodial” stage, its habit of forming aggregates of trophozoites that appear to be aggregated and enmeshed in po- lysaccharide appears similar to the pseudoplasmodial stage in dictyostelids.
In addition, characteristics of some members of the phylum Myxozoa (WEISER 1955; J. WEISER, in LEE et al.
1985) can also be compared to P. carinii. Members of
this phylum are generally parasites of fish and have or- ganized polar capsules and or filaments, but some mem- bers appear similar to P. carinii cytologically with its diffuse, multisized trophozoite form and highly orga- nized cyst (for example Chloromyxum leydigi and Tet- ractinomyxon intermedium). Nothing has been de- scribed at the molecular level for these organisms.
Figure 4, A and B, is ordered so that the protists are grouped on the top. It is worth noting that some of the protist actin genes are as diverged from each other as much
as they are from plants or animals or fungi as determined by measuring the distance lines of the tree. In addition, actins from three species classified in the fungi fell within the protist grouping. P. carinii’s actin, while grouped with the fungal actins in this study, is more closely related to the vertebrate actins
(as
determined by measuring the distance lines on the tree) than to two other ascomycetous actins (S. cer&aeand C . albicans).
P. carinii’s long parasitic as-sociation with mammalian hosts has possibly made an im- pact on its genome, adding constraints to its evolutionary rate. It is clear that more fungal and protist actins, espe- cially those from organisms with structural similarities to P. can’nii and histories of host-parasite relationships, need to be examined before definitive conclusions can be made. Finally, an analysis of additional P. carinii genes should be performed. If every P. c a ~ n i i gene falls out into a similar tree, a classification with the fungi for this parasite should be contemplated.
We are grateful to LOUISE GORTON, WYNELLA BRAKE and SUSAN JONES
750 Leah D. Fletcher et al.
thank MICHAEL DYKSTRA for helpful discussions on protist taxonomy. This work was supported by National Institutes of Health grant 1 UO1 AI33363.
LITERATURE CITED
CHAGAS, C., 1909 Nova tripanozomiaze humana. Mem. Inst. Oswaldo Cruz 1: 159-218.
CUSHION, M., J. RUFFOLO and P. WALZER, 1988 Analysis of the devel- opmental stages of Pneumocystis carinii in vitro. Lab. Invest. 58:
DEVEREUX, J., P. HAEBERLI and 0. SMITHIES, 1984 A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:
EDMAN, J., J. KOVACS, H. MASUR, D. SANTI, H. ELWOOD et al., 1988 Ri-
bosomal RNA sequence shows Pneumocystis carinii to be a mem- ber of the fungi. Nature 334 519-522.
EDMAN, J. C., U. EDMAN, M. CAo, B. LUNDGREN, J. A. KOVACS et al.,
1989 Isolation and expression of the Pneumocystis carinii di- hydrofolate reductase gene. Proc. Natl. Acad. sci. USA 8 6
EDMAN, U., J. C. EDMAN, B. LUNDGREN and D. V. SANTI, 1989 Isolation and expression of the Pneumocystis carinii thymidylate synthase gene. Proc. Natl. Acad. Sci. USA 86: 6503-6507.
FIDEL, S., J. H. DOONAN and N. R. MORRIS, 1988 Aspergillus nidulans
contains a single actin gene which has unique intron locations and encodes a yactin. Gene 7 0 283-293.
FLETCHER, L. D., L. C. BERGER, S. A. PEEL, R. S. BARIC, R. R. TIDWELL et al.,
1993 Isolation and identification of six Pneumocystis carinii
genes utilizing codon bias. Gene 129: 167-174.
KOST, T., N. THEODORAKIS and S. HUGHES, 1983 The nucleotide se- quence of the chick+ actin gene. Nucleic Acids Res ll: 8287- 8301.
LEE, J. L., S. H. HUTNER and E. C. Bovee, 1985 An Illustrated Guide
to the Protozoa. Society of Protozoologists, Lawrence, Kans. LEES-MILLER, J. P., G. HENRY and D. M. HELFMAN, 1992 Identification
of act2: an essential gene in the fission yeast Schizosaccharomyces pombe that encodes a protein related to actin. Proc. Natl. Acad. Sci. USA 89: 80-83.
LEVINE, N. D., J. 0. CORLISS, F. E. G. Cox, G. DEROUX, J. GRAIN et al.,
1980 A newly revised classification of the Protozoa. J. Protozool.
LI, W. H., C. I. Wu and C. C. Luo, 1985 A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2: 150-174.
MARCHUK, D., M. DRUMM, A. SAULINO and F. S. COLLINS, 1991 Con- struction of T-vectors, a rapid and general system for direct clon- ing of unmodified PCR products. Nucleic Acids Res. 19: 1154.
MCLEAN, B. G., S. R. HUANG, E. C. MCKINNEY and R. B. MEAGHER,
1990 Plants contain highly divergent actin isovariants. Cell Motil. Cytoskeleton 17: 276-290.
MERTINS, P., and D. Gwwrrz, 1987 A single actin gene in the fission yeast S chizosaccharomyces pombe: nucleotide sequences and tran- scripts formed in homologous and heterologous yeast. Nucleic Acids Res. 15: 7369-7379.
Nc, R., and J. AEELSON, 1980 Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 77:
324-331.
387-395.
8625-8629.
27: 37-58.
3912-3916.
OLIVE, L. S., 1975 T h e Mycetozoans. Academic Press, New York. POLLARD, T. D., 1990 Actin. Curr. Opin. Cell Biol. 2 33-40.
POLLARD, T. D. J. A,, and COOPER, 1986 Actin and actin-binding pro- teins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 5 5 987-1035.
ROMANS, P., and R. A. FIRTEL, 1985 Organization of the actin multi- gene family of Dictyostelium discoideum and analysis ofvariability in the protein coding regions. J. Mol. Biol. 186 321-335.
SAITOU, N., and M. NEI, 1987 The neighbor-joining method: a new method for reconstructuring phylogenetic trees. Mol. Biol. Evol.
SAMBROOK, J., E. FRITSCH and T. MANIATIS, 1989 Molecular Cloning:
A Laboratory M a n u a l , Ed. 2. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
SCHWOB, E., and R. P. MARTIN, 1992 New yeast actin-like gene re- quired late in the cell cycle. Nature 355: 179-182.
SOUTHERN, E. M., 1975 Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98: 503.
STOSSEL, T. P., 1984 Contribution of actin to the structure of the cytoplasmic matrix. J. Cell Biol. 99: 15s-21s.
STRINGER, S. L., J. R. STRINGER, M. A. BLASE, P. D. WALZER and M. T. CUSHION, 1989 Pneumocystis carinii: sequence from ribosomal RNA implies a close relationship with fungi. Exp. Parasitol. 6 8
SWOFFORD, D., 1991 PAUP: Phylogenetic Analysis Using Parsimony (version 3.1). Computer program distributed by the Illinois Natural History Survey, Champaign, Ill.
TAYLOR, J. W., and B. H. BOWMAN, 1993 MicroCorrespondence: Pneu- mocystis carinii and the ustomycetous red yeast fungi. Mol.
Microbiol. 8: 425-427.
TIDWELL, R. R., S. K. JONES, J. D. GERATZ, K A. OHEMENG, C. A. BELL et al.,
1990 Development of pentamidine analogues as new agents for the treatment of Pneumocystis carinii pneumonia. Ann. N.Y. Acad. Sci. 616: 421-441.
VANDEKERCKHOM, J., AND K WEBER, 1984 Chordate muscle actins dif- fer distinctly from invertebrate muscle actins. J. Mol. Biol. 179
391-413.
WAKEFIELD, A. E., S. E. PETERS, S. BANERJI, P. D. BRIDGE, G . S. HALL et al.,
1992 Pneumocystis carinii shows DNA homology with the us-
tomycetous red yeast fungi. Mol. Microbiol. 6 1903-1911.
WATANABE, J., H. HORI, K TANABE and Y. NMURA, 1989 Phyloge- netic association of Pneumocystis carinii with the ‘Rhizopoda/ Myxomycota/Zygomycota’ indicated by comparison of 5 s ribc- soma1 RNA sequences. Mol. Biochem. Parasitol. 32: 163-167.
WEISER, J., 1955 A new classification of the Schizogregarina.
J. Protozool. 2: 6-12.
WESSELING, J., M. SMITS and J. SCHOENMAKERS, 1988a Extremely di- verged actin proteins in Plasmodium falciparum. Mol. Biochem. Parasitol. 30: 143-154.
WESSELING, J. G., J. M. DEREE, T. PONNUDURAI, M. A. SMITS and J. G . G.
SCHOENMAKERS, 1988b Nucleotide sequence and deduced amino
acid sequence of a Plasmodium falciparum actin gene. Mol. Bb-
chem. Parasitol. 27: 313-320.
WESSELING, J., P. SNIPERS, P. VANSOMEREN, J. JANSEN, M. SMln et al.,
1989 Stage-Specific expression and genomic organization of the actin genes of the malaria parasite Plasmodium fakiparum.
Mol. Biochem. Parasitol. 3 5 167-176.
4: 406-425.
450-461.