0095-1137/97/$04.0010
Copyright © 1997, American Society for Microbiology
Clonal Analysis and Identification of Epidemic Strains
of Methicillin-Resistant
Staphylococcus aureus
by
Antibiotyping and Determination of Protein A
Gene and Coagulase Gene Polymorphisms
A. HOEFNAGELS-SCHUERMANS,1,2W. E. PEETERMANS,2M. J. STRUELENS,3S. VAN LIERDE,4
ANDJ. VAN ELDERE1*
Infectious Diseases Research Group, Department of Microbiology and Immunology,1
Department of Internal Medicine,2and Department of Paediatrics,4Rega Institute
for Medical Research, Katholieke Universiteit Leuven, 3000 Leuven, and Unite´ d’Epidemiologie, Laboratoire de Microbiologie, Hoˆpital Erasme,
Universite´ Libre de Bruxelles, Bruxelles,3Belgium
Received 27 January 1997/Returned for modification 11 June 1997/Accepted 10 July 1997
Forty-three methicillin-resistantStaphylococcus aureus(MRSA) isolates with known genetic and
epidemio-logical relatedness and different degrees of transmission were analyzed by antibiotyping, protein A gene poly-morphism analysis, and coagulase gene polypoly-morphism analysis. The three typing systems were evaluated for their performance and convenience to define clones and to discriminate between epidemic MRSA (EMRSA)
and sporadic MRSA (SMRSA). Antibiotyping andAluI restriction fragment length polymorphism analysis of
the coagulase gene were able to define clones in the same way as DNA macrorestriction analysis (SmaI).
How-ever, both techniques presented disadvantages, making neither of them useful as a single typing method. Pro-tein A gene polymorphism analysis appeared to be of no value for clonal analysis. None of the three typing methods was able to differentiate between EMRSA and SMRSA.
Outbreaks of hospital-acquired infections due to methicillin-resistant Staphylococcus aureus (MRSA) are being reported with increasing frequency, challenging clinicians and infection control teams throughout the world (3, 26, 29, 33). Some MRSA strains, the so-called epidemic MRSA (EMRSA) strains, are able to rapidly spread among patients. Once intro-duced into an institution these EMRSA strains are difficult to control and to eradicate. Other MRSA strains, however, lack the capacity to spread extensively.
In the 1950s it was already clear that certain strains ofS. au-reuswere able to cause outbreaks in hospitals (35). In the early 1980s a single strain of MRSA called EMRSA 1 caused out-breaks in several hospitals in England and Wales (5, 30). Ten years later Kerr et al. reported the existence of 13 other multi-hospital EMRSA strains, which affected more than one hospi-tal and were characterized by phage typing, antibiotic suscep-tibility tests, and selected biochemical tests (12). At about the same time, Struelens and coworkers demonstrated, by DNA macrorestriction and pulsed-field gel electrophoresis, that the same type 1a MRSA strain caused concurrent epidemics in at least three different Belgian hospitals (24).
Our own experience at Leuven University Hospital confirms the existence of both EMRSA and nonepidemic, or sporadic, MRSA (SMRSA) (10). EMRSA strains are mainly acquired by intensive care unit (ICU) patients, and these strains are able to spread. SMRSA strains, on the other hand, are mostly import-ed into the hospital by long-term-care residents and do not spread.
Discrimination between EMRSA and SMRSA strains is im-portant in terms of a hospital infection control policy. Detec-tion of a discriminative marker would allow a more selective implementation of infection control measures in order to pre-vent dissemination of MRSA strains within the hospital. Sev-eral attempts to identify such a marker were made. Roberts and Gaston found a lower level of protein A expression and a higher level of coagulase expression in EMRSA strains than in sporadic strains (21). Frenay and coworkers tried to discrimi-nate between EMRSA and SMRSA strains on the basis of protein A gene polymorphism (8). They reported that strains with more than seven repeats in the X region of the protein A tended to be epidemic, while the presence of seven or fewer repeats was indicative of a nonepidemic MRSA strain. Van Wamel and coworkers tried to differentiate between EMRSA and SMRSA based upon differences in binding to extracellular matrix proteins such as fibronectin, vitronectin, collagen, Fc fragments of immunoglobulin G, and fibrinogen (32). EMRSA was found to bind significantly less fibrinogen and Fc frag-ments of immunoglobulin G. Goh and coworkers successfully identified the outbreak MRSA strains in their institution by PCR amplification of the variable region of the coagulase gene followed byAluI restriction and analysis of restriction fragment length polymorphism (RFLP) (9).
The aim of our study was to evaluate three different epide-miological typing systems—antibiotyping, protein A gene poly-morphism analysis, and coagulase gene RFLP analysis—for their performance and convenience in differentiating be-tween EMRSA and SMRSA. These three techniques were also evaluated for their capacity to define clones compared to two reference typing systems, phage typing and DNA macrorestriction analysis. We chose these three systems be-cause of their ease of use and accessibility for every routine laboratory.
* Corresponding author. Mailing address: Rega Institute for Medi-cal Research, Katholieke Universiteit Leuven, Minderbroedersstraat 10, 3000 Leuven, Belgium. Phone: 32 16 33 73 72. Fax: 32 16 33 73 40. E-mail: johan.vaneldere@rega.kuleuven.ac.be.
2514
on May 15, 2020 by guest
http://jcm.asm.org/
MATERIALS AND METHODS
Bacterial isolates.The bacterial isolates were obtained from clinical specimens of hospitalized patients and residents of long-term-care facilities. Sampling was done during two 1-week periods in October 1993 and April 1994 and two 1-week periods in July 1996 and September 1996.
Standard microbiological methods for identification ofS. aureus included Gram staining, growth on mannitol salt agar (Difco, Detroit, Mich.), and cata-lase, DNase, and coagulase testing. AllS. aureusisolates for which the MIC of oxacillin was 4mg/ml or more in Mueller-Hinton agar (Difco) with 2% NaCl were classified as MRSA strains. Methicillin resistance was confirmed by the presence of themecAgene, determined by PCR as described previously (17).
Strains were stored in a brain heart infusion broth-glycerol mixture (3:1, vol/vol) at270°C until further analysis.
Two coagulase-negative isolates were included as negative controls for the PCR testing of the coagulase gene polymorphism.
Reference typing systems.Clonal analysis was performed by phage typing and DNA macrorestriction (SmaI) followed by computer-assisted normalization and clustering and as described below.
(i) Phage typing.Phage typing was done as described by Blair and Williams with the group III international set of phages, kindly provided by C. Godard of the ‘Pasteur Instituut van Brabant’, Brussels, Belgium (2). The isolates that were susceptible to lysis by a single phage only or not susceptible at all at the routine test dilution were tested with 100 times the routine test dilution of the phage suspension. Bacterial isolates were considered to belong to different phage types if they were lysed by at least two different phages. The phage types were defined by the numbers of the phages to which they were susceptible. Weak reactions were also taken into account to define the final phage pattern (2).
(ii) DNA macrorestriction analysis and pulsed-field gel electrophoresis. Iso-lation of chromosomal DNA was performed as described by Struelens et al. (24). For each isolate, 5 ml of an overnight culture in brain heart infusion was pelleted by centrifugation at 15,5003gfor 3 min. After being washed in EET buffer (100 mM EDTA, EGTA, 10 mM Tris hydrochloride [pH 8]), the bacteria were resuspended in EET buffer and the suspension was adjusted to a density of 109 CFU/ml by cell count and densitometry (optical density at 650 nm50.6). The bacterial suspension was subsequently mixed with an equal volume of 2% (wt/ vol) low-melting-temperature agarose (Incert Agarose FMC; Tech Gen, Val-lensback Strand, Denmark) in EET buffer and left to solidify. Solid agarose plugs were then incubated in EET buffer plus 0.1 mg of lysostaphin (Ambicin L; Applied Microbiology, New York, N.Y.) per ml for 4 h at 30°C to lyse the bacteria. Agarose plugs were subsequently incubated in EET buffer containing 1% (wt/vol) sodium dodecyl sulfate and 1 mg of pronase per ml for 18 h at 37°C. Protein digestion products were removed by washing the plugs six times for 30 min in 10 mM Tris–1 mM EDTA. The plugs were analyzed immediately or stored in the 10 mM Tris–1 mM EDTA buffer at 4°C until further analysis.
Macrorestriction of genomic DNA was done with 10 U ofSmaI (Gibco BRL, Life Technologies, Gaithersburg, Md.) for 4 h at 30°C. Separation of the restric-tion fragments was carried out via pulsed-field gel electrophoresis in a CHEF MAPPER system (Bio-Rad Laboratories, Hercules, Calif.) in 1% (wt/vol) chro-mosomal-grade agarose gels. Electrophoresis was performed at 200 V in a buffer containing 44.5 mM Tris hydrochloride, 44.5 mM boric acid, and 1 mM EDTA (pH 8) with alternating pulses at a 120° angle in a 5- to 15-s pulse time gradient for 10 h and a 15- to 45-s pulse time gradient for the next 12 h. A temperature of 14°C was maintained throughout the run. A DNA polymer (Pharmacia, Uppsala, Sweden) and DNA from S. aureusNTCC 8325 cut bySmaI were included as molecular size markers (20). After electrophoresis, gels were stained with ethidium bromide, rinsed, and photographed under UV light.
Definition of EMRSA versus SMRSA and analysis of strain relatedness.A strain was classified as EMRSA if isolates of the strain showed the capacity to spread among patients. This implied that for a given period these isolates were acquired by many patients during their hospitalization in the same ward (time and space clustering) and that these isolates were all genetically related.
Strains were considered SMRSA if they were recovered from only one patient. If more isolates with the same genotype were recovered from different patients, transmission between these patients could be excluded, since they were never hospitalized on the same ward and/or during the same period. This implied that these isolates did not spread.
Genetic relatedness was based on the results of DNA macrorestriction analysis and defined as advocated by Struelens et al. and Tenover et al. (25, 28).
Isolates were classified as genetically closely related to the EMRSA if their macrorestriction pattern differed from EMRSA pattern by changes consistent with a single genetic event, which typically results in a two- to three-band difference. These isolates showed a Dice coefficient of correlation of 80% or more with EMRSA. Isolates were classified as possibly genetically related to the EMRSA if their macrorestriction pattern differed from the EMRSA pattern by changes consistent with two independent genetic events, i.e., a four- to six-band difference, and if they had a Dice coefficient of correlation of 68 to 79%. Isolates were classified as genetically unrelated to EMRSA if they showed a six-band or greater difference and a Dice coefficient of correlation of 60% or less.
Banding patterns were compared visually by two independent observers and by calculating the Dice coefficients of correlation (number of shared fragments3
23100/total number of fragments in the two samples) with the Gel Compar software (Applied Maths, Kortrijk, Belgium) (6).
Epidemiologic relatedness was based on data obtained by analysis of patients charts. The isolates were considered epidemiologically related to EMRSA when they were recovered during the same time frame or from the same area during the patients’ hospital stay, according to the guidelines proposed by Tenover et al. (28).
Evaluation of typing systems.The typing systems were evaluated (i) for the capacity to define clones and (ii) for the ability to discriminate between EMRSA and SMRSA. In addition we also assessed the typing ability, reproducibility, and discriminatory power (11) of each typing system as defined by Maslow and Mulligan (15).
(i) Antibiotic susceptibility testing.Susceptibility testing was performed by agar dilution, according to National Committee for Clinical Laboratory Stan-dards guidelines (19). Antibiotics tested were penicillin, ampicillin, oxacillin, erythromycin, clindamycin, trimethoprim-sulfamethoxazole, gentamicin, tobra-mycin, amikacin, netilmicin, nitrofurantoin, imipenem, fusidic acid, rifampin, novobiocin, ofloxacin, and vancomycin. Isolates showing an intermediate level of susceptibility were classified as resistant. Bacterial isolates were considered to belong to different antibiotypes if at least one difference was observed (24).
(ii) PCR amplification and restriction analysis of the staphylococcal protein A gene.PCR amplified the highly polymorphic sequence of the X region of the protein A gene, which is composed of a variable number of 24-bp repeats. Strains were analyzed as described previously by Frenay et al. with some modifications (8). An isolated bacterial colony on a blood agar plate was suspended in 1 ml of sterile water and centrifuged for 1 min at 15,5003g. The pellet was resuspended in 200ml of Instagene matrix (Bio-Rad Laboratories) and incubated at 56°C for 15 to 30 min. The sample was heated to 100°C for 8 min and centrifuged at 15,5003gfor 3 min. The forward primer 59TGTAAAACGACGGCCAGTGC TAAAAAGCTAAACGATGC and the reverse primer CAGGAAACAGCTAT GACCCCACCAAATACAGTTGTACC were used. Ten microliter of cell lysate was added to a PCR mixture containing 1mM of each primer, 4ml of 10-fold-concentrated PCR buffer, 200mM of each deoxynucleoside triphosphate, and 1 U ofTaqpolymerase (Perkin-Elmer, Roch Molecular Systems, Inc. Branchburg, N.J.) in a final volume of 40ml of deionized water. Each sample was subjected to 25 cycles, each of 1 min at 95°C, 1 min at 60°C, and 1 min at 72°C (Perkin-Elmer thermal cycler 480).
Ten microliters of the PCR product was cut overnight with 5 U ofRsaI (Pharmacia) according to the manufacturer’s recommendations. Both the PCR products and the restriction fragments were detected by electrophoresis through a 3% Nusieve GTG agarose gel (FMC Bioproducts, Rockland, Maine) in the presence of ethidium bromide and photographed under UV illumination.
(iii) PCR amplification and restriction analysis of the staphylococcal coagu-lase gene.Strains were analysed as described by Goh et al. with some modifica-tions (9). Bacterial DNA was obtained as described above. The 39-end region of the coagulase gene was amplified with two primers: COAG 2 (59CGAGACCA AGATTCAACAAG 39) and COAG 3 (59 AAAGAAAACCACTCACATCA 39). Ten microliters of cell lysate was added to a PCR mixture containing 1mM each primer, 4ml of 10-fold concentrated PCR buffer, 200mM each deoxynucleo-side triphosphate, and 1 U ofTaqpolymerase (Perkin-Elmer) in a final volume of 40ml of deionized water. Each sample was subjected to 40 cycles, each of 30 s at 95°C, 2 min at 55°C, and 4 min at 72°C. Ten microliters of the PCR product was cut overnight with 10 U ofAluI (Boehringer Mannheim, GmbH, Mannheim, Germany) according to the manufacturer’s recommendations. Both the PCR products and the restriction digest fragments were detected by electrophoresis through a 2% Nusieve GTG agarose gel (FMC Bioproducts) in the presence of ethidium bromide and photographed under UV illumination. TheAluI RFLP patterns were given numeric codes as defined by Chow (4). This code consists of as many digits as there are bands in the RFLP pattern. Starting with the smallest fragment, each digit gives the multiple of 81 bp found in the band that it correlates with.
RESULTS
Assignment of the isolates to categories of genetic and epi-demiologic relatedness through DNA macrorestriction
analy-sis and phage typing.Eighty-seven isolates were collected from
83 patients. For 81 patients, only one MRSA strain was iso-lated. For two patients, four and two isolates with different DNA macrorestriction types were recovered on different oc-casions. With DNA macrorestriction analysis, these 87 isolates were divided into 25 different DNA macrorestriction types (Table 1). Phage typing showed 10 different phage patterns. Of the 87 isolates, 21 (24%) were not typeable by the group III international set of phages.
DNA macrorestriction types 1/I and 1/II were found to be the predominant types in terms of frequency of isolation in our hospital. They were isolated from 25 and 31 patients,
on May 15, 2020 by guest
http://jcm.asm.org/
tively. Twenty of the 25 isolates (80%) with DNA tion type 1/I and 23 of the 31 isolates (74%) with macrorestric-tion type 1/II were collected from patients in the ICU. We found that these isolates were acquired by these patients dur-ing their stay in the ICU (10). For 9 of the remaindur-ing 13 macrorestriction type 1/I and 1/II isolates, the isolate was prob-ably acquired in the ICU but was isolated from the patient on the ward following ICU stay. From these observations we con-clude that these types 1/I and 1/II have the capacity to spread between patients. Isolates of macrorestriction type 1/I were compared to the Belgian epidemic strain and were all indistin-guishable (24). The dominant DNA macrorestriction types 1/I and 1/II differed by only one band in the 360-kb region. Ac-cording to the criteria proposed by Tenover and Struelens (25, 28) this indicates that these two macrorestriction types are closely related. Interestingly, phage typing suggested a genetic distinction between macrorestriction type 1/I and 1/II. A sig-nificant correlation between macrorestriction types 1/I and 1/II and the phage types A and B was found. Of the 16 phage-typeable macrorestriction type 1/I isolates, 15 (94%) showed phage pattern B (Table 1) and 1 showed phage pattern F. Of
the 29 phage-typeable isolates of macrorestriction type 1/II, 20 (69%) revealed phage pattern A and only 3 had phage pattern B. The remaining macrorestriction type 1/II isolates belonged to types H, D, E, and J or were not typeable.
Phage pattern A is the classical epidemic pattern found in the EMRSA strains that were dominant in Belgian hospitals in the late 1980s. Pattern B is the recent epidemic phage pattern found in the EMRSA strains that became predominant in Belgian hospitals in 1993 (34).
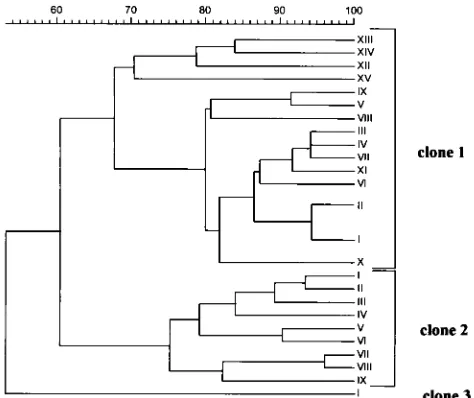
[image:3.612.60.556.83.415.2]Ten isolates belonging to nine DNA macrorestriction types were isolated from 10 patients. These nine DNA macrorestric-tion types—1/III, 1/IV, 1/V, 1/VI, 1/VII, 1/VIII, 1/IX, 1/X, and 1/XI—were genetically closely related to macrorestriction types 1/I and 1/II, with a Dice coefficient of correlation of 80 to 86%. They showed phage pattern B in eight cases, phage pat-tern G in one case, and phage patpat-tern H in one case. These isolates were all considered epidemiologically related to elec-trotypes 1/I and 1/II because of their time and space clustering. However, these isolates were classified as sporadic rather than epidemic because they were not frequently isolated and be-cause spread to other patients could not be demonstrated. TABLE 1. Genetic and epidemiologic relatedness of MRSA strains as determined by three typing systems
PFGE clonal groupa
Genetic
relatednessa Subtype
Epidemiologic relatednessb
Epidemic character
Phage pattern(s)b,c (no. of isolates)
No. of patients
Antibiotype(s)d (no. of isolatese)
RFLP result(s) for:
Protein A genef
(no. of isolatese
)
Coagulase geneg
(no. of isolatese
)
1 Closely related I Yes Yes B (15), F (1), NT (9) 25 a (5) 11 (5) 1,2,5 (5)
II Yes Yes A (20), B (3), D (1), E (1),
H (3), J (1), NT (2)
31 a (2), e (3), f (5) 11 (9), 10 (1) 1,2,5 (10)
III Yes No B 1 d (1) 11 (1) 1,2,5 (1)
IV Yes No B 1 c (1) 11 (1) 1,2,5 (1)
V Yes No B (1), G (1) 2 c (1), e (1) 11 (1), 10 (1) 1,2,5 (2)
VI Yes No B 1 a (1) 11 (1) 1,2,5 (1)
VII Yes No B 1 a (1) 11 (1) 1,2,5 (1)
VIII Yes No B 1 j (1) 10 (1) 1,2,5 (1)
IX Yes No B 1 a (1) 11 (1) 1,2,5 (1)
X Yes No B 1 b (1) 10 (1) 1,2,5 (1)
XI Yes No H 1 a (1) 11 (1) 1,2,5 (1)
Possibly related XII No No A (1), B (1) 2 c (1) 10 (1) 1,2,5 (1)
XIII No No B (1), D (1) 2 h (1) 11 (1) 1,2,5 (1)
XIV No No A 1 g (1) 10 (1) 1,2,5 (1)
XV No No A 1 f (1) 11 (1) 1,2,5 (1)
2 Not related I No No G (2), NT (2) 4 i (4) 9 (2), 10 (2) 1,3,6 (4)
II No No NT 1 i (1) 10 (1) 1,3,6 (1)
III No No NT 1 i (1) 9 (1) 1,3,6 (1)
IV No No NT 1 i (1) 10 (1) 1,3,6 (1)
V No No B2 1 i (1) 9 (1) 1,3,6 (1)
VI No No NT (2) 2 i (1) 10 (1) 1,3,6 (1)
VII No No NT 1 i (1) ? (1) 1,3,6 (1)
VIII No No NT 1 i (1) 10 (1) 1,3,6 (1)
IX No No NT (2) 2 i (2) 10 (2) 1,3,6 (2)
3 Not related I No No I 1 k (1) 10 (1) 10 (1)
aPFGE, pulsed-field gel electrophoresis. bRelatedness to type 1/I.
cPhage patterns: A, 42E-47-54-75-77-83A-84-85/811; B, 47-54-75-77-84-85; B2, 75-77-84-85; D, 47-54-75-85; E, 47-54-77-83A; F, 42E-54-75-77-83A-85; G, 77-84; H, 42E-47-54-75-83A-85/811; I, 6-42E-47-53-54-75-77-84-85-81; J, 54; NT, not typeable.
dFor complete antibiotic susceptibility profiles, see Table 2.
eThe total number of isolates is 43; for antibiotyping and protein A gene and coagulase gene polymorphism analysis, only a random sample of the 87 isolates was
tested.
fData are numbers of 24-bp repeats.
gData are multiples of 81 bp (each digit corresponds to one band).
on May 15, 2020 by guest
http://jcm.asm.org/
DNA macrorestriction types 1/XII, 1/XIII, 1/XIV, and 1/XV were classified as genetically possibly related to types 1/I and 1/II, with a Dice correlation coefficient of 68%. Three isolates showed phage pattern A, two showed phage pattern B, and one showed phage pattern D (Table 1). They were classified as not epidemiologically related to the epidemic strains because of the lack of space clustering: all strains originated from different wards. In addition, they were not epidemic in terms of fre-quency of isolation. The six isolates belonging to the four macrorestriction types were found in six patients, and trans-mission between patients could be excluded.
All macrorestriction types already mentioned were desig-nated clonal group 1 (Fig. 1). Fifteen isolates from 15 patients representing 10 macrorestriction types (2/I, 2/II, 2/III, 2/IV, 2/V, 2/VI, 2/VII, 2/VIII, 2/IX, and 3/I) (Table 1) were not genetically or epidemiologically related to macrorestriction types 1/I or 1/II. They showed a Dice coefficient of correlation
of 60% or less with macrorestriction types 1/I and 1/II (Fig. 1). Four isolates belonged to macrorestriction type 2/I and were recovered from four geriatric patients, but direct transmission between these patients could be excluded. Two of these four patients were hospitalized in two separate geriatric wards and the other two patients were residents of two distant long-term-care facilities. Except for the single macrorestriction type 3/I isolate, all these macrorestriction types were genetically closely related (less than a four-band difference or a Dice coefficient of correlation of 75 to 95%). Consequently, macrorestriction types 2/I, 2/II, 2/III, 2/IV, 2/V, 2/VI, 2/VII, 2/VIII, and 2/IX were considered to belong to the same clonal group, group 2. The unique isolate of macrorestriction type 3/I was considered to constitute clonal group 3 (Fig. 1). All isolates of clones 2 and 3 were classified as SMRSA.
Antibiotyping and protein A and coagulase gene
polymor-phism analysis as epidemiological typing systems.Forty-three
of the 87 isolates were randomly selected for further analysis: 29 isolates were from clone 1, 13 were from clone 2, and 1 was from clone 3 (Table 1). Based on the criteria of genetic relat-edness, a distinction was made within clone 1 between isolates that were genetically closely related and genetically possibly related to macrorestriction type 1/I (28). On the basis of the epidemiological data and the criteria described above, 15 iso-lates of clone 1 were classified as EMRSA and 28 isoiso-lates originating from clones 1, 2, and 3 were considered SMRSA. Each isolate was subjected to antibiotyping and protein A and coagulase gene polymorphism analysis in order to determine to what extent each method could identify clonal relatedness and predict epidemic behavior.
(i) Antibiotyping. Eleven distinct antibiotypes were
identi-fied (Table 2). Of 15 EMRSA isolates, 7 showed antibiotype a, 5 showed antibiotype f, and 3 showed antibiotype e. Of the 10 genetically closely related isolates that belonged to clone 1, 4 showed antibiotype a and two showed antibiotype c. Antibio-types b, d, e, and j were found in only 1 isolate each. All four genetically possibly related isolates that belonged to clone 1 showed different antibiotypes: c, f, g, and h. No single antibiotic or combination of antibiotics could discriminate between the different subtypes of clone 1. All 29 isolates belonging to clone 1 were resistant to ofloxacin, erythromycin, clindamycin, gen-FIG. 1. Dendrogram based on the Dice coefficient of pattern similarity,
[image:4.612.60.297.69.268.2]ob-tained by DNA macrorestriction analysis (SmaI).
TABLE 2. Antibiotic susceptibilities of 43 MRSA isolates
Drug
Resultafor isolates of antibiotype:
a (11)b b (1) c (3) d (1) e (4) f (6) g (1) h (1) i (13) j (1) k (1)
Penicillin R R R R R R R R R R R
Ampicillin R R R R R R R R R R R
Oxacillin R R R R R R R R R R R
Ofloxacin R R R R R R R R R R R
Erythromycin R R R R R R R R S R R
Clindamycin R R R R R R R R S R S
Trimethoprim-sulfamethoxazole S S S S S S S S S S S
Gentamicin R R R R R R R R R R R
Tobramycin R R R R R R R R R R R
Amikacin R S R S R S R R S S R
Netilmicin R R R R S S R R S S R
Imipenem R R R R R R R R S R R
Fusidic acid S S S S S S S R S S S
Nitrofurantoin S S S S S S R S S S S
Novobiocin S S S S S S S S S S S
Rifampin R S S R R R S S S S S
Yancomycin S S S S S S S S S S S
aR, resistant; S, susceptible.
bNumbers in parentheses are numbers of isolates.
on May 15, 2020 by guest
http://jcm.asm.org/
tamicin, and tobramycin. Of these 29 isolates, 76% were resis-tant to rifampin and 69% were resisresis-tant to amikacin.
All 13 isolates of clone 2 showed antibiotype i. Antibiotype i isolates were all resistant to gentamicin and ofloxacin but susceptible to erythromycin, amikacin, clindamycin, and ri-fampin. Therefore, erythromycin and clindamycin were clonal markers for clone 1 and 2 strains.
The unique macrorestriction type 3/I isolate representing clone 3 showed antibiotype k. The clone 3 strain was resistant to ofloxacin, gentamicin, amikacin, and erythromycin but sus-ceptible to clindamycin.
All of the 15 EMRSA strains were resistant to at least 10 of the 14 antibiotics tested. Of the 28 SMRSA isolates, on the other hand, 14 were resistant to fewer than 10 of the 14 anti-biotics tested. All isolates were typeable by antibiotyping. The reproducibility was good, and the interstrain discriminatory index was high (0.84).
(ii) Protein A gene polymorphism analysis.Amplification by
PCR of the highly polymorphic sequence of the X region of the protein A gene and restriction byRsaI generated a constant fragment of 214 bp, a 35-bp fragment not visible on the gel, and a third fragment containing a variable number (from 9 to 11) of 24-bp repeats (Table 1). Of the 15 EMRSA isolates of clone 1, 14 showed 11 repeats and 1 had 10 repeats. Of the 10 ge-netically closely related isolates of clone 1, 7 had 11 repeats and 3 had 10 repeats. Of the four genetically possibly related isolates of clone 1, two showed 11 repeats and two showed 10 repeats. For clone 2, we found no isolate with 11 repeats, but 8 isolates of 13 showed 10 repeats and 4 showed 9 repeats. For one particular isolate of clone 2 (macrorestriction type 2/VII), repeated analysis failed to produce a banding pattern. The single isolate of clone 3 showed 10 repeats. On the basis of these results, discrimination between isolates belonging to dif-ferent clones was not possible. In addition, no distinction could be made between isolates with different spreading behaviors within clone 1. When considering the ability of the results of protein A gene polymorphism analysis to differentiate between EMRSA and SMRSA strains, we found the following: of the 28 SMRSA isolates, 9 showed 11 repeats, 14 had 10 repeats, and 4 had 9 repeats; of the 15 EMRSA isolates, 14 showed 11 repeats and 1 had 10 repeats.
The typing ability of protein A gene polymorphism analysis was 0.97, and the reproducibility was 100%. The interstrain discriminatory index was low (0.58).
(iii) Coagulase gene polymorphism analysis.AluI restriction
of the PCR amplification product of the 39 region of the co-agulase gene generated multiple DNA fragments. Two coagu-lase-negative staphylococci served as negative controls and produced no DNA products upon PCR amplification. PCR amplification and restriction of the coagulase gene of the 43 MRSA isolates generated three different patterns (Table 1). All 29 isolates of clone 1 showed the RFLP pattern 1,2,5. All 13 isolates of clone 2 showed the pattern 1,3,6. The unique clone 3 isolate showed only one fragment, corresponding to 10 repeats. On the basis of these RFLP patterns, isolates could be assigned to the correct clone—1, 2, or 3—as defined by mac-rorestriction analysis.
Prediction of the spreading behavior (EMRSA versus SMRSA), on the other hand, was not possible. All EMRSA isolates revealed the same 1,2,5 RFLP pattern, but for the SMRSA both RFLP patterns were equally represented (14 isolates with a 1,2,5 pattern, 13 isolates with a 1,3,6 pattern, and 1 isolate with a single fragment).
All strains were typeable; the technique was 100% repro-ducible, but the discriminatory index was low (0.46).
DISCUSSION
In this study we evaluated antibiotyping and protein A gene and coagulase gene polymorphism analysis for their capacity to define clonal relatedness and to detect markers of epidemicity. It is clear that rapid and reliable discrimination between MRSA with possible epidemic behavior (EMRSA) and MRSA without the tendency to spread (SMRSA) is of crucial impor-tance in any attempt to prevent MRSA outbreaks.
The clonal relatedness of the set of isolates used in this study was first determined on the basis of DNA macrorestriction analysis because this has become the new “gold standard” for epidemiological typing (1). The guidelines proposed by Teno-ver and the European Study Group on Epidemiological Mark-ers have resolved the issue of standardization and interpreta-tion of genomic typing patterns (25, 28). Three separate clones named 1, 2, and 3 were identified. Combined with analysis of patient data, the results of DNA macrorestriction analysis al-lowed division of clone 1 isolates into EMRSA and two groups of genetically related nonepidemic isolates (28). Macrorestric-tion type 1/I is the Belgian EMRSA type defined by Struelens and coworkers (24). Although type 1/II was genetically very closely related to type 1/I, we considered it a different EMRSA type because of its different phage pattern. Most EMRSA isolates had either phage pattern A or B, previously identified as the Belgian epidemic patterns (34). In spite of its limita-tions, we performed phage typing to complement the results of macrorestriction analysis because it has long been considered the gold standard (1, 16, 31).
Antibiotyping allowed a clear distinction between clone 1 and 2 isolates on the basis of their susceptibility to erythromy-cin and clindamyerythromy-cin and was therefore found to be an accept-able clonal marker for this set of isolates. Nevertheless, it lacks the discriminatory power of DNA macrorestriction analysis, and the inherent instability of the plasmids makes it unreliable over extended periods. Antibiotyping is, however, the simplest epidemiologic typing method, the results are easy to interpret, and minimal laboratory skills and equipment are required.
Protein A gene polymorphism analysis reportedly discrimi-nates between EMRSA and SMRSA but has not been claimed as a method for defining clonality (8). A match between the numbers of 24-bp repeats and the clones defined by DNA macrorestriction analysis could not be demonstrated. Never-theless, as strains became genetically more distant from the EMRSA, the percent 11-repeat isolates decreased and the percents 10- and 9-repeat isolates increased. Interestingly, one isolate was repeatedly not typeable, suggesting absence of the X region of the protein A gene, which encodes the cell wall binding domain. This could imply production of exclusively extracellular protein A or the absence of the protein A gene (7).
Coagulase gene RFLP patterns matched perfectly with the clonal delineation defined by DNA macrorestriction analysis. All strains of clone 1 had the pattern 1,2,5 whereas all strains of clone 2 showed the pattern 1,3,6. Coagulase gene RFLP analysis has been reported as a new and attractive typing method for clinical laboratories (9, 13, 18, 22). It requires only small quantities of crude DNA, and isolates can be compared easily by both the number of PCR products and the sizes of theirAluI restriction fragments. The low discriminatory power makes it less suitable as a single typing method, but this could be overcome by the use of other enzymes to expand the num-ber of bands (27).
The second aim of our study was to evaluate the capacity of these three techniques to discriminate between EMRSA and SMRSA. Extensive epidemiologic data were obtained which,
on May 15, 2020 by guest
http://jcm.asm.org/
in conjunction with DNA macrorestriction analysis, made the identification of epidemic versus sporadic phenotypes unequiv-ocal. The frequency of isolation and the observation of trans-mission clearly differentiated between epidemic and nonepi-demic strains (10). A clear distinction needs to be made between genetic relatedness, epidemiologic relatedness, and epidemic behavior. Some epidemiologically unrelated isolates may have similar or indistinguishable genotypes, particularly if there is limited genetic diversity within a species or subtype. This is especially true for most strains of MRSA, which are derived from a small number of ancestral clones (14, 27). In our study it became clear that isolates can be both genetically and epidemiologically closely related to an epidemic strain without showing a tendency to spread. Possibly these strains arose from long-standing circulating epidemic strains through loss of epidemic markers. Typing studies performed on isolates for which epidemiologic information is not available may pro-duce misleading information. Therefore, strain typing does not replace epidemiologic data. Rather, these two data sets should be developed independently but analyzed together to deter-mine strain relatedness and to anticipate the capacity of the strain to cause major hospital outbreaks.
Antibiotyping was unable to predict the epidemic behavior of an isolate, although EMRSA tended to be multiresistant. Multidrug resistance may contribute to the ability to survive in the hospital environment but cannot be considered the only factor that contributes to the spread of EMRSA in hospitals. Discrimination between EMRSA and SMRSA on the basis of protein A gene polymorphism with a cutoff value of seven 24-bp repeats in the X region was proposed by Frenay and coworkers (8). A longer X region would allow a more favorable exposure of the Fc binding regions at the cell surface, facili-tating colonization and infection of the skin or other not-yet-identified sites that may be important for epidemic spread. In our study, all isolates showed 9 to 11 repeats, independently of their epidemic behavior. Our observations are in concordance with the report of Senna and coworkers, who also were not able to validate the cutoff value of seven repeats (23). It has been suggested that multiple loci of the chromosome may contribute to the epidemic character of a strain. The number of 24-bp repeats in the X region of the protein A gene might be only one of these markers.
For the coagulase gene RFLP analysis, too, the patterns were not a marker of epidemiological relatedness and were not able to differentiate between isolates with and without an ep-idemic character within the same clone. Moreover, the two RFLP patterns were equally represented within the SMRSA group.
Interestingly, four isolates from four different patients were indistinguishable by DNA macrorestriction analysis and clus-tered in macrorestriction type 2/I. Whether these strains rep-resent a new emerging clone with epidemic behavior is not clear at this moment. No epidemiological relatedness could be demonstrated for these four isolates.
In conclusion, antibiotyping is valuable, especially in routine laboratories, as a first-line screening method to determine strain relatedness. It may allow quick and early recognition of a previously defined epidemic strain in a particular hospital setting. Coagulase gene polymorphism analysis provides easy and early analysis of clonal relatedness of isolates. Protein A gene polymorphism analysis was not a valuable method of clonal analysis. None of the three typing methods was able to make a distinction between EMRSA and SMRSA. The epi-demic factor has not been characterized. Undoubtedly, multi-ple factors contribute to the ability of staphylococci to spread in hospitals.
ACKNOWLEDGMENTS
We thank Anthony W. Chow from the University of British Colom-bia, Vancouver, Canada, for stimulating discussions and support of this study. We thank Jan Verhaegen from the Laboratory of Bacteriology, University Hospitals Leuven, and Marc Lontie from the M.C.H., Lab-oratory of Microbiology, Leuven, for kindly providing MRSA strains from long-term-care facilities. We also thank Rita Merckx for excellent technical assistance.
REFERENCES
1.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and M. Miller.1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of
Staphylococcus aureus. J. Clin. Microbiol.33:551–555.
2.Blair, J. E., and R. E. O. Williams.1961. Phage typing of staphylococci. Bull. W. H. O.24:771–784.
3.Brumfitt, W., and J. Hamilton-Miller.1989. Methicillin-resistant Staphylo-coccus aureus. N. Engl. J. Med.320:1188–1196.
4.Chow, A. W.1995. Personal communication.
5.Cookson, B. D., and I. Phillips.1988. Epidemic methicillin-resistant Staph-ylococcus aureus. J. Antimicrob. Chemother.21:57–65.
6.Dice, L. R.1945. Measures of the amount of ecological association between species. Ecology26:297–302.
7.Forsgren, A., V. Ghetie, R. Lindmark, and J. Sjo¨quist.1983. Protein A and its exploitation, p. 429–480.InS. F. Easmon and C. Adlam (ed.), Staphylo-cocci and staphylococcal infections. Academic Press Ltd., London, England. 8.Frenay, H. M. E., J. P. G. Theelen, L. M. Schouls, C. M. J. E. Vandenbro-ucke-Grauls, J. Verhoef, W. J. van Leeuwen, and F. R. Mooi.1994. Discrim-ination of epidemic and nonepidemic methicillin-resistantStaphylococcus aureusstrains on the basis of protein A gene polymorphism. J. Clin. Micro-biol.32:846–847.
9.Goh, S., S. K. Byrne, J. L. Zhang, and A. W. Chow.1992. Molecular typing ofStaphylococcus aureus on the basis of coagulase gene polymorphism. J. Clin. Microbiol.30:1642–1645.
10. Hoefnagels-Schuermans, A., A. Borremans, W. E. Peetermans, S. Van Lierde, G. Reybrouck, and J. Van Eldere.1997. Origin and transmission of methicillin-resistantStaphylococcus aureusin an endemic situation: differ-ences between geriatric and intensive-care patients. J. Hosp. Infect.36:209– 222.
11. Hunter, P. R., and M. A. Gaston.1988. Numerical index of the discrimina-tory ability of typing systems: an application of Simpson’s index of diversity. J. Clin. Microbiol.26:2465–2466.
12. Kerr, S., G. E. Kerr, C. A. Mackintosh, and R. R. Marples.1990. A survey of methicillin-resistantStaphylococcus aureusaffecting patients in England and Wales. J. Hosp. Infect.16:35–48.
13. Kobayashi, N., K. Taniguchi, K. Kojima, S. Urasawa, N. Uehara, Y. Omize, Y. Kishi, A. Yagihashi, and I. Kurokawa.1995. Analysis of methicillin-resistant and methicillin-susceptibleStaphylococcus aureusby a molecular typing method based on coagulase gene polymorphisms. Epidemiol. Infect.
115:419–426.
14. Kreiswirth, B., J. Kornblum, D. Arbeit, W. Eisner, J. N. Maslow, A. Mc-Geere, D. E. Low, and R. P. Novick.1993. Evidence for a clonal origin of methicillin-resistance inStaphylococcus aureus. Science259:227–230. 15. Maslow, J., and M. E. Mulligan.1996. Epidemiologic typing systems. Infect.
Control Hosp. Epidemiol.17:595–604.
16. Mulligan, M. E., and R. D. Arbeit.1991. Epidemiologic and clinical utility of typing systems for differentiating among strains of methicillin resistant Staph-ylococcus aureus. Infect. Control Hosp. Epidemiol.12:20–28.
17. Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabee.1991. Identification of methicillin-resistant strains of staphylo-cocci by polymerase chain reaction. J. Clin. Microbiol.29:2240–2244. 18. Nada, T., S. Ichiyama, Y. Osada, M. Ohta, K. Shimokata, N. Kato, and N.
Nakashima.1996. Comparison of DNA fingerprinting by PFGE and PCR-RFLP of the coagulase gene to distinguish MRSA isolates. J. Hosp. Infect.
32:305–317.
19. National Committee for Clinical Laboratory Standards.1994. Performance standards for antimicrobial disk susceptibility testing, 5th ed. Approved standard M2-A5. National Committee for Clinical Laboratory Standards, Villanova, Pa.
20. Pattee, P. A.1989. Genetic and physical mapping of the chromosome of
Staphylococcus aureusNCTC 8325, p. 163–169.InK. Drlica and M. Riley (ed.), The bacterial chromosome. American Society for Microbiology, Wash-ington, D.C.
21. Roberts, J. I. S., and M. A. Gaston.1987. Protein A and coagulase expression in epidemic and non-epidemicStaphylococcus aureus. J. Clin. Pathol.40:837– 840.
22. Schwarzkopf, A., and H. Karch.1994. Genetic variation inStaphylococcus aureuscoagulase genes. Potential and limits for use as epidemiological marker. J. Clin. Microbiol.32:2407–2412.
23. Senna, J., E. Chachaty, P. Saulnier, E. Espaze, N. Barbier, B. Leclercq, H. Richet, and A. Andremont.1996. Molecular characterization of sporadic and
on May 15, 2020 by guest
http://jcm.asm.org/
epidemic methicillin-resistantStaphylococcus aureusstrains in 3 French hos-pitals, abstr. 285, p. 260.InProgram and abstracts of the 8th International Symposium on Staphylococci and Staphylococcal Infections. Socie´te´ Fran-c¸aise de Microbiologie, Aix-Les-Bains, France.
24. Struelens, M. J., A. Deplano, C. Godard, N. Maes, and E. Serruys.1992. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J. Clin. Microbiol.30:2599– 2605.
25. Struelens, M. J., and the Members of the ESGEM and ESCMID.1996. Consensus guidelines for appropriate use and evaluation of microbial epi-demiologic typing systems. Clin. Microbiol. Infect.2:2–11.
26. Struelens, M. J., O. Ronveaux, B. Jans, R. Mertens, and GDEPIH.1996. Methicillin-resistantStaphylococcus aureusepidemiology and control in Bel-gian hospitals, 1991 to 1995. Infect. Control Hosp. Epidemiol.17:503–508. 27. Tenover, F. C., R. Arbeit, G. Archer, J. Biddle, S. Byrne, R. Goering, G.
Hancock, G. A. He´bert, B. Hill, R. Hollis, W. R. Jarvis, B. Kreiswirth, W. Eisner, J. Maslow, L. K. McDougal, J. M. Miller, M. Mulligan, and M. A. Pfaller.1994. Comparison of traditional and molecular methods of typing isolates ofStaphylococcus aureus. J. Clin. Microbiol.32:407–415. 28. Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. R. Murray,
D. H. Persing, and B. Swaminathan.1995. Interpreting chromosomal DNA
restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol.33:2233–2239.
29. Thompson, R. L., and R. P. Wenzel.1982. International recognition of methicillin-resistantStaphylococcus aureus. Ann. Intern. Med.97:925–926. 30. Townsend, D. E., N. Ashdown, S. Bolton, J. Bradley, G. Duckworth, E. C.
Moorhouse, and W. B. Grubb.1987. The international spread of methicillin-resistantStaphylococcus aureus. J. Hosp. Infect.9:60–71.
31. van Belkum, A., R. Bax, P. Peerbooms, W. H. F. Goessens, N. van Leeuwen, and W. G. V. Quint.1993. Comparison of phage typing and DNA finger-printing by polymerase chain reaction for discriminating methicillin-resistant
Staphylococcus aureusstrains. J. Clin. Microbiol.31:798–803.
32. Van Wamel, W. J. B., A. C. Fluit, T. Wadstro¨m, H. van Dijk, J. Verhoef, and C. M. J. E. Vandenbroucke-Grauls.1995. Phenotypic characterization of epidemic versus sporadic strains of methicillin-resistantStaphylococcus au-reus. J. Clin. Microbiol.33:1769–1774.
33. Voss, A., D. Milatovic, C. Wallrauch-Schwarz, V. T. Rosdahl, and I. Braveny.
1994. Methicillin resistantStaphylococcus aureusin Europe. Eur. J. Clin. Microbiol. Infect Dis.13:50–55.
34. Widemauwe, C., C. Godard, R. Vanhoof, E. Van Bossuyt, and E. Hannecart-Pokorni.1996. Changes in major populations of methicillin-resistant Staph-ylococcus aureusin Belgium. J. Hosp. Infect.34:197–203.
35. Williams, R. E. O.1959. Epidemic staphylococci. Lancetii:190–195.