organic papers
o672
Hino and Mizuguchi C38H24N4O42C7H8O doi:10.1107/S1600536805004447 Acta Cryst.(2005). E61, o672–o674 Acta Crystallographica Section E
Structure Reports Online
ISSN 1600-5368
N
,
N
000-Bis[2-(4-pyridyl)ethyl]perylene-3,4:9,10-bis(dicarboximide)
m
-cresol disolvate
Kazuyuki Hino and Jin Mizuguchi*
Department of Applied Physics, Graduate School of Engineering, Yokohama National University, Tokiwadai 79-5, Hodogaya-ku, Yokohama 240-8501, Japan
Correspondence e-mail: mizu-j@ynu.ac.jp
Key indicators
Single-crystal X-ray study T= 93 K
Mean(C–C) = 0.008 A˚ Rfactor = 0.100 wRfactor = 0.118
Data-to-parameter ratio = 12.0
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
#2005 International Union of Crystallography Printed in Great Britain – all rights reserved
The title compound, C38H24N4O42C7H8O, is a 1:2 complex of a pyridylethylperylene derivative, EPY, with m-cresol. The EPY molecule has a centre of symmetry and the pyridylethyl groups are attached to the perylene–imide skeleton in atrans
fashion. The EPY molecules are stacked along theaaxis with a slip angle of about 47.
Comment
Perylene compounds are industrially important pigments, covering a variety of shades from red via maroon to black (Herbst & Hunger, 1993). N,N0
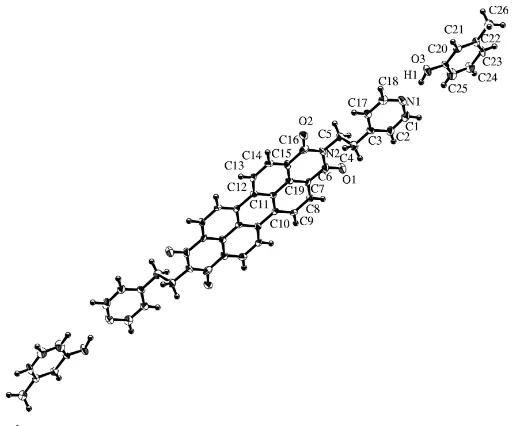
[image:1.610.204.473.535.655.2]-Bis[2-(4-pyridyl)ethyl]-perylene-3,4:9,10-bis(dicarboximide) is a pyridylethyl deriva-tive, here abbreviated to EPY. It has a similar structure to that of the phenylethyl derivative, here abbreviated to EPH and also known as pigment black 31. The only difference between EPY and EPH is the pyridyl or phenyl ring. Nevertheless, the colours are strikingly different. EPY (Mizuguchi & Tojo, 2002) is vivid red, while two crystal modifications of EPH (Ha¨dicke & Graser, 1986; Mizuguchi, 1998) are black. In these two modifications of EPH, the phenylethyl groups are attached to the perylene imide skeleton in a trans fashion, while the pyridylethyl groups in EPY arecis(Mizuguchi & Tojo, 2002). Because of this, our attention has been focused on the preparation of the trans form of EPY, which is expected to show a black colour. We have recently isolated a black crystal of the trans form from a 1:1 mixed solvent of phenol and ethanol, EPY2(phenol) (Mizuguchi & Hino, 2005a). The present paper deals with another complex of thetransform, EPY2(m-cresol), (I), recrystallized fromm-cresol and which is red–black.
Fig. 1 shows the structure of (I), in which one molecule of EPY crystallizes with two m-cresol molecules. The EPY molecule has a centre of symmetry and the two pyridylethyl groups are arranged in a trans fashion (Ci symmetry), in
marked contrast to the previous cis form (C2 symmetry; Mizuguchi & Tojo, 2002). The pyridyl rings deviate from the perylene imide skeleton by 5.2 (2).
There is an intermolecular O—H N hydrogen bond (Table 2) between the m-cresol molecule and the pyridyl group of EPY. Them-cresol molecule is twisted by 18.6 (2)
with respect to the pyridyl ring. The colour of complex (I) is reddish-black, as expected (Mizuguchi & Hino, 2005b).
Fig. 2 shows the projection of the crystal structure along the
a axis. The EPY molecules form columns along the a axis, while there are two neighbouring columns composed of
m-cresol molecules. The polar m-cresol molecules of each column are arranged so as to cancel their dipole moments to reduce the electrostatic energy. Within a column, the EPY molecules are stacked with a slip angle of about 47, which is
defined, in a side view of two stacked molecules, as the slipped angle of the upper molecule relative to the lower one along the long molecular axis.
Experimental
EPY was prepared by the reaction of perylene tetracarboxylic dianhydride (10 g) with aminoethylpyridine (8.8 g) at 403 K in water (30 ml) for 5 h. The product was filtered and the red cake was then refluxed for 10 min inN,N0-dimethylformamide. Single crystals of a
reddish-black colour (transform) were grown from solution inm -cresol, whereas red crystals (cisform) were obtained from solution in nitrobenzene. The use of a protic solvent such asm-cresol was the key to the growth of black crystals of thetransform. Since the reddish-black crystal of (I) was found to include solvent molecules, X-ray intensity data were collected at 93 K. Obtaining single crystals from the vapour phase was also attempted but without success, leading to the decomposition of EPY to give a perylene imide derivative known as pigment violet 29.
Crystal data
C38H24N4O42C7H8O
Mr= 816.88
Monoclinic,P21=c
a= 4.903 (6) A˚
b= 29.26 (4) A˚
c= 13.96 (2) A˚
= 97.65 (6)
V= 1985 (4) A˚3
Z= 2
Dx= 1.367 Mg m
3
CuKradiation Cell parameters from 4038
reflections
= 3.0–65.6 = 0.73 mm1
T= 93.2 K Needle, red
0.400.100.10 mm
Data collection
Rigaku R-AXIS RAPID imaging plate diffractometer
!scans
Absorption correction: multi-scan (ABSCOR; Higashi, 1995)
Tmin= 0.618,Tmax= 0.930
3443 measured reflections
3363 independent reflections 1335 reflections withF2> 2(F2)
Rint= 0.147 max= 68.2
h= 0!5
k= 0!35
l=16!16
Refinement
Refinement onF2 R[F2> 2(F2)] = 0.100
wR(F2) = 0.118
S= 1.00 3363 reflections 280 parameters
H-atom parameters constrained
w= 1/[2
(Fo2) + {0.025[Max(Fo2,0) +
2Fc2]/3}2]
(/)max< 0.001
max= 1.08 e A˚ 3
min=0.90 e A˚3
[image:2.610.42.302.69.282.2] [image:2.610.47.295.329.530.2]Extinction correction: none
Table 1
Selected bond lengths (A˚ ).
O1—C6 1.223 (7) O2—C16 1.233 (7) N2—C5 1.493 (7) N2—C6 1.405 (7) N2—C16 1.412 (7) C6—C7 1.469 (8) C7—C8 1.370 (7) C7—C19 1.406 (8) C8—C9 1.410 (7) C9—C10 1.384 (7)
C10—C11 1.437 (7) C10—C12i
1.485 (7) C11—C12 1.429 (8) C11—C19 1.425 (7) C12—C13 1.402 (7) C13—C14 1.393 (7) C14—C15 1.387 (8) C15—C16 1.465 (8) C15—C19 1.399 (8) C17—C18 1.387 (7)
[image:2.610.315.566.548.636.2]Symmetry codes: (i)xþ1;yþ1;zþ2.
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
O3—H1 N1ii
1.00 1.80 2.761 (5) 161
organic papers
Acta Cryst.(2005). E61, o672–o674 Hino and Mizuguchi C
38H24N4O42C7H8O
o673
Figure 1A view of the molecular conformation of (I), showing 50% probability displacement ellipsoids. Unlabelled atoms are related to labelled atoms by 1x, 1y, 2z.
Figure 2
[image:2.610.312.567.698.729.2]The H atom attached to the O atom was found in a difference-density map and its positional parameters were fixed withUiso(H) =
1.1Ueq(O). All other H atoms were positioned geometrically and
included in a riding-model approximation, with C—H = 0.95 A˚ and withUiso(H) = 1.2Ueq(C). The maximum residual density is located
1.25 A˚ from atom C5.
Data collection: PROCESS-AUTO (Rigaku, 1998); cell refine-ment: PROCESS-AUTO; data reduction: TEXSAN (Molecular Structure Corporation, 2001); program(s) used to solve structure:
SYSTEM90(Houet al., 1994); program(s) used to refine structure:
TEXSAN; molecular graphics: ORTEPIII (Burnett & Johnson, 1996); software used to prepare material for publication:TEXSAN.
References
Burnett, M. N. & Johnson, C. K. (1996).ORTEPIII. Report ORNL-6895. Oak Ridge National Laboratory, Tennessee, USA.
Ha¨dicke, E. & Graser, F. (1986).Acta Cryst.C42, 189–195.
Herbst, W. & Hunger, K. (1993).Industrial Organic Pigments, pp. 467-475. Weinheim: VCH.
Higashi, T.(1995).ABSCOR. Rigaku Corporation, Tokyo, Japan. Hou, Y., Gao, M., Li, L. & Hou, P. (1994).Acta Cryst.A50, 748–753. Mizuguchi, J. (1998).Acta Cryst.C54, 1479–1481.
Mizuguchi, J. & Hino, K. (2005a).Acta Cryst.E61, o672–o674 Mizuguchi, J. & Hino, K. (2005b).Dyes Pigments. Submitted.
Mizuguchi, J. & Tojo, K. (2002).Z. Kristallogr. New Cryst. Struct.217, 247–248. Molecular Structure Corporation (2001).TEXSAN. Version 1.11. MSC, 9009
New Trails Drive, The Woodlands, TX 77381-5209, USA.
Rigaku (1998).PROCESS-AUTO. Rigaku Corporation, Tokyo, Japan.
organic papers
o674
Hino and Mizuguchi Csupporting information
sup-1 Acta Cryst. (2005). E61, o672–o674
supporting information
Acta Cryst. (2005). E61, o672–o674 [https://doi.org/10.1107/S1600536805004447]
N
,
N
′-Bis[2-(4-pyridyl)ethyl]perylene-3,4:9,10-bis(dicarboximide)
m
-cresol
disolvate
Kazuyuki Hino and Jin Mizuguchi
(I)
Crystal data
C38H24N4O4·2C7H8O
Mr = 816.88 Monoclinic, P21/c
Hall symbol: -P 2ybc
a = 4.903 (6) Å
b = 29.26 (4) Å
c = 13.96 (2) Å
β = 97.65 (6)°
V = 1985 (4) Å3
Z = 2
F(000) = 856.0
Dx = 1.367 Mg m−3
Cu Kα radiation, λ = 1.5419 Å Cell parameters from 4038 reflections
θ = 3.0–65.6°
µ = 0.73 mm−1
T = 93 K Needle, red
0.40 × 0.10 × 0.10 mm
Data collection
Rigaku R-AXIS RAPID imaging plate diffractometer
Detector resolution: 10.00 pixels mm-1
24 frames, δω = 30° scans Absorption correction: multi-scan
(ABSCOR; Higashi, 1995)
Tmin = 0.618, Tmax = 0.930
3443 measured reflections
3363 independent reflections 1335 reflections with F2 > 2σ(F2)
Rint = 0.147
θmax = 68.2°
h = 0→5
k = 0→35
l = −16→16
Refinement
Refinement on F2
R[F2 > 2σ(F2)] = 0.100
wR(F2) = 0.118
S = 1.00 3363 reflections 280 parameters
H-atom parameters constrained
w = 1/[σ2(F
o2) + {0.025[Max(Fo2,0) + 2Fc2]/3}2]
(Δ/σ)max = 0.0002
Δρmax = 1.08 e Å−3
Δρmin = −0.90 e Å−3
Special details
Refinement. Refinement using reflections with F2 > -3.0 σ(F2). The weighted R-factor (wR) and goodness of fit (S) are
based on F2. R-factor (gt) are based on F. The threshold expression of F2 > 2.0 σ(F2) is used only for calculating R-factor
(gt).
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
supporting information
sup-2 Acta Cryst. (2005). E61, o672–o674
O2 −0.5191 (7) 0.4996 (1) 0.6876 (3) 0.036 (1)
O3 −1.5941 (7) 0.6440 (1) 0.1952 (2) 0.037 (1)
N1 −1.1697 (9) 0.6368 (2) 0.3457 (3) 0.033 (2)
N2 −0.3847 (9) 0.5714 (1) 0.7349 (3) 0.024 (2)
C1 −0.992 (1) 0.6661 (2) 0.3919 (4) 0.037 (2)
C2 −0.777 (1) 0.6536 (2) 0.4621 (4) 0.033 (2)
C3 −0.730 (1) 0.6081 (2) 0.4846 (4) 0.027 (2)
C4 −0.501 (1) 0.5930 (2) 0.5611 (4) 0.038 (2)
C5 −0.607 (1) 0.5898 (2) 0.6613 (4) 0.032 (2)
C6 −0.205 (1) 0.6024 (2) 0.7880 (4) 0.032 (2)
C7 0.019 (1) 0.5836 (2) 0.8572 (4) 0.023 (2)
C8 0.199 (1) 0.6124 (2) 0.9109 (4) 0.024 (2)
C9 0.4173 (10) 0.5956 (2) 0.9777 (4) 0.021 (2)
C10 0.459 (1) 0.5491 (2) 0.9903 (4) 0.023 (2)
C11 0.275 (1) 0.5185 (2) 0.9328 (4) 0.020 (2)
C12 0.312 (1) 0.4701 (2) 0.9415 (4) 0.022 (2)
C13 0.128 (1) 0.4422 (2) 0.8824 (4) 0.022 (2)
C14 −0.090 (1) 0.4598 (2) 0.8189 (4) 0.027 (2)
C15 −0.127 (1) 0.5067 (2) 0.8100 (4) 0.025 (2)
C16 −0.355 (1) 0.5235 (2) 0.7405 (4) 0.032 (2)
C17 −0.914 (1) 0.5772 (2) 0.4345 (4) 0.028 (2)
C18 −1.128 (1) 0.5928 (2) 0.3672 (4) 0.032 (2)
C19 0.054 (1) 0.5360 (2) 0.8662 (4) 0.023 (2)
C20 −1.721 (1) 0.6858 (2) 0.1791 (4) 0.025 (2)
C21 −1.944 (1) 0.6882 (2) 0.1046 (4) 0.025 (2)
C22 −2.082 (1) 0.7288 (2) 0.0829 (4) 0.026 (2)
C23 −2.002 (1) 0.7670 (2) 0.1378 (4) 0.035 (2)
C24 −1.778 (1) 0.7645 (2) 0.2119 (4) 0.038 (2)
C25 −1.637 (1) 0.7241 (2) 0.2320 (4) 0.041 (2)
C26 −2.321 (1) 0.7306 (2) 0.0014 (4) 0.041 (2)
H1 −1.4375 0.6487 0.2475 0.0395*
H2 −1.0141 0.6975 0.3762 0.0449*
H3 −0.6623 0.6765 0.4949 0.0396*
H4 −0.4366 0.5640 0.5440 0.0453*
H5 −0.3560 0.6147 0.5645 0.0453*
H6 −0.7616 0.5700 0.6564 0.0384*
H7 −0.6585 0.6194 0.6804 0.0384*
H8 0.1754 0.6444 0.9035 0.0281*
H9 0.5384 0.6165 1.0141 0.0260*
H10 0.1522 0.4100 0.8858 0.0267*
H11 −0.2142 0.4396 0.7818 0.0314*
H12 −0.8942 0.5454 0.4472 0.0335*
H13 −1.2494 0.5708 0.3345 0.0379*
H14 −2.0019 0.6614 0.0692 0.0301*
H15 −2.0974 0.7950 0.1251 0.0419*
H16 −1.7229 0.7911 0.2486 0.0449*
H17 −1.4842 0.7228 0.2815 0.0498*
supporting information
sup-3 Acta Cryst. (2005). E61, o672–o674
H19 −2.2751 0.7131 −0.0517 0.0485*
H20 −2.4799 0.7184 0.0232 0.0485*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
O1 0.042 (3) 0.030 (3) 0.034 (3) 0.006 (2) −0.004 (2) 0.003 (2)
O2 0.030 (3) 0.035 (3) 0.038 (3) −0.000 (2) −0.006 (2) 0.003 (2)
O3 0.045 (3) 0.034 (3) 0.028 (3) 0.007 (2) −0.013 (2) 0.003 (2)
N1 0.033 (4) 0.039 (3) 0.025 (3) 0.010 (3) −0.002 (3) 0.002 (3)
N2 0.020 (3) 0.026 (3) 0.025 (3) 0.007 (3) −0.001 (2) 0.002 (3)
C1 0.049 (5) 0.020 (4) 0.043 (4) 0.004 (3) 0.010 (4) 0.003 (3)
C2 0.036 (4) 0.026 (4) 0.036 (4) −0.003 (3) −0.000 (3) 0.005 (3)
C3 0.019 (4) 0.042 (4) 0.020 (4) 0.004 (3) 0.005 (3) 0.007 (3)
C4 0.034 (4) 0.051 (4) 0.028 (4) 0.001 (3) 0.006 (3) 0.008 (3)
C5 0.029 (4) 0.037 (4) 0.031 (4) 0.004 (3) 0.007 (3) 0.007 (3)
C6 0.036 (5) 0.023 (4) 0.037 (5) 0.008 (3) 0.009 (4) 0.009 (3)
C7 0.028 (4) 0.017 (4) 0.025 (4) −0.005 (3) 0.004 (3) 0.006 (3)
C8 0.022 (4) 0.020 (4) 0.029 (4) 0.007 (3) 0.006 (3) 0.003 (3)
C9 0.016 (4) 0.023 (4) 0.025 (4) −0.006 (3) 0.004 (3) −0.002 (3)
C10 0.022 (4) 0.026 (4) 0.022 (4) −0.003 (3) 0.007 (3) 0.007 (3)
C11 0.016 (4) 0.026 (4) 0.019 (4) 0.003 (3) 0.010 (3) −0.000 (3)
C12 0.020 (4) 0.024 (4) 0.023 (4) 0.003 (3) 0.004 (3) 0.003 (3)
C13 0.028 (4) 0.017 (3) 0.023 (4) −0.001 (3) 0.005 (3) 0.000 (3)
C14 0.023 (4) 0.034 (4) 0.022 (4) −0.005 (3) 0.004 (3) −0.003 (3)
C15 0.030 (4) 0.032 (4) 0.013 (3) 0.004 (3) 0.001 (3) 0.004 (3)
C16 0.034 (5) 0.037 (4) 0.028 (4) −0.006 (4) 0.017 (3) −0.002 (4)
C17 0.028 (4) 0.028 (4) 0.028 (4) 0.004 (3) 0.005 (3) −0.003 (3)
C18 0.038 (5) 0.033 (4) 0.028 (4) 0.004 (3) 0.010 (3) −0.003 (3)
C19 0.022 (4) 0.027 (4) 0.019 (4) −0.001 (3) −0.000 (3) −0.002 (3)
C20 0.019 (4) 0.031 (4) 0.023 (4) 0.003 (3) −0.003 (3) 0.005 (3)
C21 0.028 (4) 0.022 (3) 0.024 (4) −0.004 (3) 0.004 (3) −0.007 (3)
C22 0.029 (4) 0.024 (4) 0.025 (4) 0.005 (3) 0.002 (3) 0.003 (3)
C23 0.042 (4) 0.027 (4) 0.036 (4) 0.020 (4) 0.002 (3) 0.006 (3)
C24 0.048 (5) 0.029 (4) 0.035 (4) −0.003 (4) −0.001 (3) −0.009 (3)
C25 0.057 (5) 0.044 (4) 0.020 (4) −0.003 (4) −0.006 (3) −0.006 (3)
C26 0.040 (4) 0.039 (4) 0.044 (4) −0.001 (3) 0.005 (3) 0.009 (4)
Geometric parameters (Å, º)
O1—C6 1.223 (7) C10—C12i 1.485 (7)
O2—C16 1.233 (7) C11—C12 1.429 (8)
O3—C20 1.378 (6) C11—C19 1.425 (7)
O3—H1 0.996 C12—C13 1.402 (7)
N1—C1 1.327 (7) C13—C14 1.393 (7)
N1—C18 1.333 (7) C13—H10 0.951
N2—C5 1.493 (7) C14—C15 1.387 (8)
supporting information
sup-4 Acta Cryst. (2005). E61, o672–o674
N2—C16 1.412 (7) C15—C16 1.465 (8)
C1—C2 1.390 (8) C15—C19 1.399 (8)
C1—H2 0.949 C17—C18 1.387 (7)
C2—C3 1.382 (8) C17—H12 0.950
C2—H3 0.951 C18—H13 0.950
C3—C4 1.507 (7) C20—C21 1.404 (7)
C3—C17 1.399 (7) C20—C25 1.377 (8)
C4—C5 1.558 (8) C21—C22 1.380 (8)
C4—H4 0.948 C21—H14 0.950
C4—H5 0.950 C22—C23 1.382 (8)
C5—H6 0.950 C22—C26 1.523 (7)
C5—H7 0.951 C23—C24 1.407 (7)
C6—C7 1.469 (8) C23—H15 0.950
C7—C8 1.370 (7) C24—C25 1.379 (8)
C7—C19 1.406 (8) C24—H16 0.950
C8—C9 1.410 (7) C25—H17 0.950
C8—H8 0.949 C26—H18 0.949
C9—C10 1.384 (7) C26—H19 0.952
C9—H9 0.950 C26—H20 0.944
C10—C11 1.437 (7)
O1···C23ii 3.543 (7) C5···C7v 3.496 (8)
O2···C18iii 3.246 (7) C6···C9v 3.434 (8)
O3···N1 2.761 (5) C6···C8v 3.592 (9)
O3···C9iv 3.356 (6) C10···C14vii 3.420 (8)
O3···C14iii 3.401 (6) C10···C13vii 3.585 (9)
O3···C18 3.431 (6) C11···C16vi 3.438 (9)
N2···C7v 3.595 (8) C11···C12vii 3.581 (9)
C1···C25vi 3.454 (9) C12···C19vii 3.428 (8)
C1···C20vi 3.460 (8) C12···C14vi 3.597 (9)
C3···C18vi 3.589 (9) C14···C18viii 3.318 (8)
C4···C18vi 3.463 (8)
C20—O3—H1 106.6 C11—C12—C13 117.5 (4)
C1—N1—C18 116.4 (4) C12—C13—C14 122.7 (5)
C5—N2—C6 118.6 (4) C12—C13—H10 118.7
C5—N2—C16 117.1 (4) C14—C13—H10 118.6
C6—N2—C16 124.0 (4) C13—C14—C15 120.0 (5)
N1—C1—C2 124.2 (5) C13—C14—H11 120.0
N1—C1—H2 117.8 C15—C14—H11 120.0
C2—C1—H2 118.0 C14—C15—C16 118.0 (5)
C1—C2—C3 120.1 (5) C14—C15—C19 119.5 (5)
C1—C2—H3 120.0 C16—C15—C19 122.5 (5)
C3—C2—H3 119.9 O2—C16—N2 118.3 (5)
C2—C3—C4 121.9 (5) O2—C16—C15 125.9 (5)
C2—C3—C17 115.6 (5) N2—C16—C15 115.8 (5)
C4—C3—C17 122.5 (5) C3—C17—C18 120.5 (5)
supporting information
sup-5 Acta Cryst. (2005). E61, o672–o674
C3—C4—H4 109.0 C18—C17—H12 119.9
C3—C4—H5 108.8 N1—C18—C17 123.3 (5)
C5—C4—H4 109.5 N1—C18—H13 118.5
C5—C4—H5 109.4 C17—C18—H13 118.2
H4—C4—H5 109.7 C7—C19—C11 119.3 (5)
N2—C5—C4 110.0 (4) C7—C19—C15 119.7 (5)
N2—C5—H6 109.5 C11—C19—C15 121.0 (5)
N2—C5—H7 109.4 O3—C20—C21 117.1 (5)
C4—C5—H6 109.3 O3—C20—C25 122.6 (4)
C4—C5—H7 109.3 C21—C20—C25 120.3 (5)
H6—C5—H7 109.4 C20—C21—C22 121.0 (5)
O1—C6—N2 118.9 (5) C20—C21—H14 119.6
O1—C6—C7 123.4 (5) C22—C21—H14 119.4
N2—C6—C7 117.7 (5) C21—C22—C23 118.7 (5)
C6—C7—C8 120.0 (5) C21—C22—C26 120.0 (5)
C6—C7—C19 120.2 (5) C23—C22—C26 121.3 (5)
C8—C7—C19 119.7 (5) C22—C23—C24 120.1 (5)
C7—C8—C9 121.8 (5) C22—C23—H15 119.9
C7—C8—H8 119.1 C24—C23—H15 120.0
C9—C8—H8 119.1 C23—C24—C25 120.9 (5)
C8—C9—C10 120.7 (4) C23—C24—H16 119.4
C8—C9—H9 119.7 C25—C24—H16 119.6
C10—C9—H9 119.6 C20—C25—C24 118.9 (5)
C9—C10—C11 118.2 (5) C20—C25—H17 120.6
C9—C10—C12i 122.6 (5) C24—C25—H17 120.5
C11—C10—C12i 119.1 (5) C22—C26—H18 109.2
C10—C11—C12 120.4 (5) C22—C26—H19 109.0
C10—C11—C19 120.3 (5) C22—C26—H20 109.4
C12—C11—C19 119.3 (5) H18—C26—H19 109.4
C10i—C12—C11 120.4 (4) H18—C26—H20 110.1
C10i—C12—C13 122.1 (5) H19—C26—H20 109.8
Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) x+2, −y+3/2, z+1/2; (iii) −x−2, −y+1, −z+1; (iv) x−2, y, z−1; (v) x−1, y, z; (vi) x+1, y, z; (vii) −x, −y+1, −z+2; (viii) −x−1, −y+1, −z+1.
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A