metal-organic papers
m172
Liet al. [Sn(C6H5)3Cl(H2O)]2C5H5N doi:10.1107/S1600536805042315 Acta Cryst.(2006). E62, m172–m174
Acta Crystallographica Section E
Structure Reports
Online
ISSN 1600-5368
Aquachlorotriphenyltin(IV) pyridine disolvate
Shun-Li Li, Jian-Fang Ma* and Ying-Ying Liu
Department of Chemistry, Northeast Normal University, Changchun 130024, People’s Republic of China
Correspondence e-mail: jianfangma@yahoo.com.cn
Key indicators
Single-crystal X-ray study
T= 293 K
Mean(C–C) = 0.004 A˚
Rfactor = 0.019
wRfactor = 0.049
Data-to-parameter ratio = 18.5
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
#2006 International Union of Crystallography Printed in Great Britain – all rights reserved
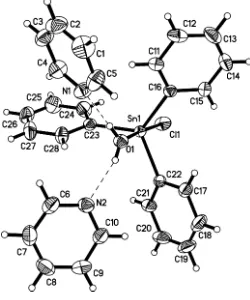
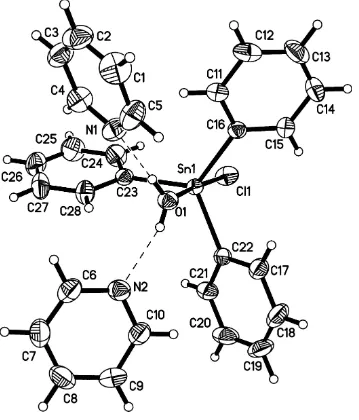
In the structure of the title mononuclear Sn complex, [Sn(C6H5)3Cl(H2O)]2C5H5N, the Sn
IV
atom is coordinated in a slightly distorted trigonal–bipyramidal geometry by three phenyl C atoms, one water molecule and one Clanion. Two pyridine molecules are O—H N hydrogen bonded to the coordinated water molecule.
Comment
In recent years, there have been many reports on the syntheses and structure determinations of various organotin(IV) compounds (e.g.Lockhartet al., 1987; Teohet al., 1997; Basuet al., 2005). These compounds have special applications, such as as PVC stabilizers, agricultural biocides, additives for anti-fouling paints, and catalysts for the production of poly-urethanes and silicones, and are potential antitumor agents (Thoonen et al., 2004). Furthermore, several structures of Ph3SnCl(H2O) cocrystallized with other molecules have been
determined, for example 3-[2-(1,10-phenanthrolyl)]-5,6-diphenyl-1,2,4-triazine (Laddet al., 1984), 3,4,7,8-tetramethyl-1,10-phenanthroline (Ng & Kumar Das, 1996), [N,N0 -bis(3-methoxysalicylidene)propane-1,3-diamine]nickel(II) (Clarke et al., 1994), di-2-pyridylketone 2-aminobenzoylhydrazone (Lanelli et al., 1995), o-phenanthroline (Ng & Kumar Das, 1996), 2,20:60,200-terpyridyl (Prasad et al., 1982), 18-crown-6
(Amini et al., 2003), 8-methoxyquinoline (Khoo et al., 2000) and di-2-pyridyl-2-thenoylhydrazone (Carcelliet al., 1995). In these structures, there is hydrogen bonding between the coordinated water molecule of Ph3SnCl(H2O) and the
cocrystallized molecule in the structure. In this paper, we report a structure in which two pyridine molecules are hydrogen bonded to Ph3SnCl(H2O).
In the molecular structure of the title compound, (I), the Sn atom is five-coordinated in a slightly distorted trigonal–
bipyramidal geometry by three C atoms of three phenyl groups in the equatorial plane, and by one Clanion and one water molecule in the axial positions (Fig. 1). The slight distortion from the ideal trigonal–bipyramidal geometry is reflected in the O1—Sn1—Cl1 angle of 175.34 (8), and the
three C—Sn—C angles of 116.54 (9), 119.84 (7) and
122.39 (7). The two pyridine molecules are connected to the coordinated water molecule through O—H O hydrogen bonds (Fig. 1 and Table 2).
Experimental
A mixture of Ph3SnCl (0.385 g, 0.1 mmol) and pyridine (0.198 g, 0.2 mmol) in 95% ethanol (13 ml) was stirred for 0.5 h. The mixture was then transferred and sealed into an 18 ml Teflon-lined autoclave, which was heated at 393 K for 89 h. After the mixture was cooled to room temperature, colorless blocks of the title complex were filtered off, washed with diethylether and dried at ambient temperature in air (yield 56% based on Sn). Analysis calculated for the title compound: C 59.88, H 4.85, N 4.99%; found: C 59.65, H 4.93, N 5.02%.
Crystal data
[Sn(C6H5)3Cl(H2O)]2C5H5N Mr= 561.66
Orthorhombic,Pna21 a= 15.492 (5) A˚ b= 15.925 (5) A˚ c= 10.885 (5) A˚ V= 2685.4 (17) A˚3 Z= 4
Dx= 1.389 Mg m 3
MoKradiation Cell parameters from 8982
reflections
= 2.2–28.2 = 1.07 mm1 T= 293 (2) K Needle, colorless 0.430.130.11 mm
Data collection
Bruker APEX CCD area-detector diffractometer
!scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996) Tmin= 0.623,Tmax= 0.882 15753 measured reflections
5668 independent reflections 5086 reflections withI> 2(I) Rint= 0.020
max= 28.4 h=14!20 k=20!20 l=10!14
Refinement
Refinement onF2 R[F2> 2(F2)] = 0.019 wR(F2) = 0.049 S= 1.04 5668 reflections 307 parameters
H atoms treated by a mixture of independent and constrained refinement
w= 1/[2
(Fo2) + (0.0281P)2] whereP= (Fo2+ 2Fc2)/3 (/)max= 0.002
max= 0.30 e A˚
3
min=0.22 e A˚
3
Extinction correction:SHELXL97 Extinction coefficient: 0.0062 (2) Absolute structure: Flack (1983),
[image:2.610.313.563.74.374.2]2299 Friedel pairs Flack parameter:0.005 (16)
Table 1
Selected geometric parameters (A˚ ,).
C16—Sn1 2.124 (2) C22—Sn1 2.127 (2) C23—Sn1 2.145 (3)
O1—Sn1 2.3469 (14) Sn1—Cl1 2.5068 (9)
C16—Sn1—C22 116.54 (9) C16—Sn1—C23 122.39 (7) C22—Sn1—C23 119.84 (7) C16—Sn1—O1 85.41 (7) C22—Sn1—O1 83.32 (7)
C23—Sn1—O1 90.01 (9) C16—Sn1—Cl1 92.74 (6) C22—Sn1—Cl1 93.72 (5) C23—Sn1—Cl1 94.59 (7) O1—Sn1—Cl1 175.34 (8)
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
O1—H1A N1 0.78 (3) 1.96 (3) 2.740 (2) 170 (3) O1—H1B N2 0.76 (3) 2.01 (3) 2.745 (2) 164 (6)
All H atoms bonded to C atoms were positioned geometrically and refined as riding atoms with C—H = 0.93 A˚ andUiso(H) = 1.2Ueq(C). The H atoms of the coordinated water molecule were located in a difference Fourier map and then refined isotropically.
Data collection:SMART(Bruker, 1997); cell refinement:SAINT
(Bruker, 1999); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics:
SHELXTL-Plus(Sheldrick, 1990); software used to prepare material for publication:SHELXL97.
We thank the National Natural Science Foundation of China (No. 20471014), the Fok Ying Tung Education Foun-dation and the Natural Science FounFoun-dation of Jilin Province (China) for support.
References
Amini, M. M., Foladi, S., Aghabozorg, H., Rare, A. D. & Ng, S. W. (2003). Chin. J. Struct. Chem.22, 77–83.
Basu, B. T. S., Rynfah, W., Rivarola, E., Pettinari, C. & Linden, A. (2005).J. Organomet. Chem.690, 1413–1421.
metal-organic papers
Acta Cryst.(2006). E62, m172–m174 Liet al. [Sn(C
6H5)3Cl(H2O)]2C5H5N
m173
Figure 1 [image:2.610.44.297.646.726.2]Bruker (1997).SMART. Version 5.622. Bruker AXS Inc., Madison, Wisconsin, USA.
Bruker (1999).SAINT. Version 6.02. Bruker AXS Inc., Madison, Wisconsin, USA.
Carcelli, M., Pelizzi, C., Pelizzi, G., Mazza, P. & Zani, F. (1995).J. Organomet. Chem.488, 55–61.
Clarke, N., Cunningham, D., Higgins, T., McArdle, P., McGinley, J. & O0
Gara, M. (1994).J. Organomet. Chem.469, 33–40.
Flack, H. D. (1983).Acta Cryst.A39, 876–881.
Khoo, L. E., Ouyang, J., Xu, Y. & Ng, S. W. (2000).Main Group Metal Chem. 23, 723–724.
Ladd, M. F. C., Povey, D. C. & Smith, F. E. (1984).J. Crystallogr. Spectrosc. Res. 14, 249–259.
Lanelli, S., Mazza, P., Orcesi, M., Pelizzi, C. & Zani, F. (1995). J. Inorg. Biochem.60, 89–108.
Lockhart, T. P., Calabrese, J. C. & Davidson, F. (1987).Organometallics,6, 2479–2483.
Ng, S. W. & Kumar Das, V. G. (1996). J. Organomet. Chem. 513, 105– 108.
Prasad, L., Lee, F. L., Le Page, Y. & Smith, F. E. (1982).Acta Cryst.B38, 259– 262.
Sheldrick, G. M. (1990).SHELXTL-Plus.Siemens Analytical X-ray Instru-ment Inc., Madison, Wisconsin, USA.
Sheldrick, G. M. (1996).SADABS. University of Go¨ttingen, Germany. Sheldrick, G. M. (1997). SHELXS97 and SHELXL97. University of
Go¨ttingen, Germany.
Teoh, S. G., Ang, S. H., Looi, E. S., Leok, C. A., Teo, S. B. & Fun, H. K. (1997). J. Organomet. Chem.527, 15–19.
Thoonen, S. H. L., Deelman, B.-J. & van Koten, G. (2004).J. Organomet. Chem.689, 2145–2157.
metal-organic papers
m174
Liet al. [Sn(Csupporting information
sup-1
Acta Cryst. (2006). E62, m172–m174
supporting information
Acta Cryst. (2006). E62, m172–m174 [doi:10.1107/S1600536805042315]
Aquachlorotriphenyltin(IV) pyridine disolvate
Shun-Li Li, Jian-Fang Ma and Ying-Ying Liu
S1. Comment
In recent years, there have been many reports on the syntheses and structure determinations of various organotin(IV)
compounds (e.g. Lockhart et al., 1987; Teoh et al., 1997; Basu et al., 2005). These compounds have special applications,
such as as PVC stabilizers, agricultural biocides, additives for antifouling paints, and catalysts for the production of
polyurethanes and silicones, and are potential antitumor agents (Thoonen et al., 2004). Furthermore, several structures of
Ph3SnCl(H2O) cocrystallized with other molecules have been determined, for example
3-[2-(1,10-phenanthrolyl)]-5,6-di-phenyl-1,2,4-triazine (Ladd et al., 1984), 3,4,7,8-tetramethyl-1,10-phenanthroline (Ng & Kumar Das, 1996), [N,N
′-bis(3-methoxysalicylidene)propane-1,3-diamine]nickel(II) (Clarke et al., 1994), di-2-pyridylketone 2-aminobenzoylhydrazone
(Lanelli et al., 1995), o-phenanthroline (Ng & Kumar Das, 1996), 2,2′:6′,2′′-terpyridyl (Prasad et al., 1982), 18-crown-6
(Amini et al., 2003), 8-methoxyquinoline (Khoo et al., 2000) and di-2-pyridyl-2-thenoylhydrazone (Carcelli et al., 1995).
In these structures, there is hydrogen bonding between the coordinated water molecule of Ph3SnCl(H2O) and the
cocrystallized molecule in the structure. In this paper, we report a structure in which two pyridine molecules are
hydrogen bonded to Ph3SnCl(H2O).
In the molecular structure of the title compound, (I), the Sn atom is five-coordinated in a slightly distorted
trigonal-bipyramidal geometry by three C atoms of three phenyl groups in the equatorial plane, and by one Cl− anion and one
water molecule in the axial positions (Fig. 1). The slight distortion from the ideal trigonal-bipyramidal geometry is
reflected in the O1—Sn1—Cl1 angle of 175.34 (8)°, and the three C—Sn—C angles of 116.54 (9), 119.84 (7) and
122.39 (7)°. The two pyridine molecules are connected to the coordinated water molecule through O—H···O hydrogen
bonds (Fig. 1 and Table 2).
S2. Experimental
A mixture of Ph3SnCl (0.385 g, 0.1 mmol) and pyridine (0.198 g, 0.2 mmol) in 95% ethanol (13 ml) was stirred for 0.5 h.
The mixture was then transferred and sealed into an 18 ml Teflon-lined autoclave, which was heated at 393 K for 89 h.
After the mixture was cooled to room temperature, colorless blocks of the title complex were filtered off, washed with
ether and dried at ambient temperature in air (yield 56% based on Sn). Analysis calculated for the title compound: C
59.88, H 4.85, N 4.99%; found: C 59.65, H 4.93, N 5.02%.
S3. Refinement
All H atoms bonded to C atoms were positioned geometrically and refined as riding atoms with C—H = 0.93 Å and
Uiso(H) = 1.2Ueq(C). The H atoms of the coordinated water molecule were located in a difference Fourier map and then
supporting information
sup-2
[image:5.610.129.488.73.493.2]Acta Cryst. (2006). E62, m172–m174 Figure 1
View of the structure of (I), showing displacement ellipsoids at the 30% probability level. Dashed lines indicate hydrogen
bonds.
Aquachlorotriphenyltin(IV) pyridine disolvate
Crystal data
[Sn(C6H5)3Cl(H2O)]·2C5H5N Mr = 561.66
Orthorhombic, Pna21
Hall symbol: P 2c -2n
a = 15.492 (5) Å
b = 15.925 (5) Å
c = 10.885 (5) Å
V = 2685.4 (17) Å3 Z = 4
F(000) = 1136
Dx = 1.389 Mg m−3
Melting point: not measured K Mo Kα radiation, λ = 0.71069 Å Cell parameters from 8982 reflections
θ = 2.2–28.2°
supporting information
sup-3
Acta Cryst. (2006). E62, m172–m174 Data collection
Bruker APEX CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
ω scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin = 0.623, Tmax = 0.882
15753 measured reflections 5668 independent reflections 5086 reflections with I > 2σ(I)
Rint = 0.020
θmax = 28.4°, θmin = 1.8° h = −14→20
k = −20→20
l = −10→14
Refinement
Refinement on F2
Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.019 wR(F2) = 0.049 S = 1.04 5668 reflections 307 parameters 1 restraint
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H atoms treated by a mixture of independent and constrained refinement
w = 1/[σ2(F
o2) + (0.0281P)2]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max = 0.002
Δρmax = 0.30 e Å−3
Δρmin = −0.22 e Å−3
Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4
Extinction coefficient: 0.0062 (2)
Absolute structure: Flack (1983), 2299 Friedels Absolute structure parameter: −0.005 (16)
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
C1 0.51354 (18) 0.82066 (19) 0.6124 (3) 0.0883 (9)
H1 0.4845 0.8540 0.6690 0.106*
C2 0.5405 (2) 0.85287 (16) 0.5045 (3) 0.0850 (8)
H2 0.5304 0.9089 0.4853 0.102*
C3 0.5825 (2) 0.80206 (19) 0.4246 (3) 0.0785 (8)
H3 0.6017 0.8227 0.3495 0.094*
C4 0.59618 (16) 0.72082 (16) 0.4556 (2) 0.0691 (6)
H4 0.6246 0.6865 0.3995 0.083*
C5 0.52989 (14) 0.73769 (15) 0.6370 (3) 0.0776 (6)
H5 0.5107 0.7155 0.7112 0.093*
C6 0.46244 (16) 0.43538 (16) 0.4866 (2) 0.0700 (6)
H6 0.4664 0.4874 0.4482 0.084*
supporting information
sup-4
Acta Cryst. (2006). E62, m172–m174
H7 0.3759 0.3896 0.3677 0.098*
C8 0.40433 (19) 0.3001 (2) 0.4905 (3) 0.0876 (9)
H8 0.3694 0.2584 0.4574 0.105*
C9 0.45174 (18) 0.28495 (17) 0.5935 (2) 0.0793 (8)
H9 0.4491 0.2331 0.6327 0.095*
C10 0.50346 (12) 0.34791 (12) 0.6381 (3) 0.0643 (5)
H10 0.5359 0.3373 0.7083 0.077*
C11 0.77724 (13) 0.68223 (14) 0.7079 (2) 0.0551 (5)
H11 0.7440 0.6884 0.6373 0.066*
C12 0.79864 (17) 0.75269 (15) 0.7782 (3) 0.0721 (7)
H12 0.7799 0.8057 0.7542 0.087*
C13 0.8473 (2) 0.7435 (2) 0.8825 (3) 0.0827 (11)
H13 0.8617 0.7905 0.9289 0.099*
C14 0.8745 (2) 0.6664 (2) 0.9188 (2) 0.0852 (9)
H14 0.9071 0.6608 0.9901 0.102*
C15 0.85403 (17) 0.59632 (15) 0.8499 (2) 0.0642 (6)
H15 0.8731 0.5438 0.8757 0.077*
C16 0.80552 (13) 0.60262 (12) 0.74313 (17) 0.0445 (4) C17 0.69499 (16) 0.39679 (14) 0.85288 (19) 0.0580 (5)
H17 0.6919 0.4498 0.8885 0.070*
C18 0.66487 (18) 0.32782 (19) 0.9169 (2) 0.0774 (8)
H18 0.6409 0.3351 0.9945 0.093*
C19 0.6699 (2) 0.2496 (2) 0.8680 (3) 0.0825 (11)
H19 0.6503 0.2035 0.9123 0.099*
C20 0.70402 (17) 0.23894 (15) 0.7529 (3) 0.0772 (8)
H20 0.7068 0.1855 0.7185 0.093*
C21 0.73442 (14) 0.30760 (14) 0.6877 (2) 0.0573 (6)
H21 0.7583 0.2997 0.6102 0.069*
C22 0.72966 (11) 0.38797 (12) 0.73637 (18) 0.0425 (4) C23 0.76955 (15) 0.49598 (10) 0.4412 (3) 0.0456 (5) C24 0.8438 (2) 0.49805 (15) 0.3697 (3) 0.0692 (8)
H24 0.8977 0.4978 0.4075 0.083*
C25 0.8386 (3) 0.50045 (16) 0.2425 (3) 0.0848 (11)
H25 0.8890 0.5026 0.1961 0.102*
C26 0.7610 (3) 0.49965 (14) 0.1849 (4) 0.0792 (10)
H26 0.7582 0.5012 0.0996 0.095*
C27 0.6874 (3) 0.49658 (15) 0.2522 (3) 0.0794 (10)
H27 0.6341 0.4951 0.2128 0.095*
C28 0.6912 (2) 0.49569 (13) 0.3795 (3) 0.0621 (7)
H28 0.6401 0.4949 0.4244 0.075*
supporting information
sup-5
Acta Cryst. (2006). E62, m172–m174 Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
C1 0.0821 (17) 0.0832 (17) 0.100 (3) 0.0289 (13) 0.0149 (17) −0.0179 (18) C2 0.0815 (18) 0.0560 (14) 0.117 (2) 0.0082 (13) −0.0076 (17) 0.0160 (15) C3 0.0849 (19) 0.0792 (18) 0.0715 (17) −0.0070 (15) 0.0072 (14) 0.0237 (15) C4 0.0689 (16) 0.0708 (15) 0.0674 (16) 0.0067 (12) 0.0105 (12) −0.0028 (12) C5 0.0716 (13) 0.0954 (16) 0.0657 (14) 0.0131 (12) 0.0106 (19) 0.019 (2) C6 0.0600 (14) 0.0763 (15) 0.0737 (16) 0.0057 (12) 0.0032 (12) 0.0255 (13) C7 0.0631 (16) 0.126 (2) 0.0551 (15) −0.0085 (16) −0.0078 (12) 0.0116 (16) C8 0.0811 (19) 0.107 (2) 0.0747 (18) −0.0416 (17) 0.0023 (15) −0.0173 (17) C9 0.0820 (17) 0.0642 (14) 0.092 (2) −0.0183 (13) 0.0126 (14) 0.0155 (12) C10 0.0534 (10) 0.0744 (13) 0.0652 (12) −0.0008 (9) −0.0013 (16) 0.020 (2) C11 0.0560 (13) 0.0454 (11) 0.0638 (14) −0.0054 (9) 0.0009 (10) −0.0016 (10) C12 0.0772 (17) 0.0462 (12) 0.093 (2) −0.0100 (11) 0.0196 (15) −0.0111 (12) C13 0.101 (2) 0.080 (2) 0.068 (2) −0.0286 (19) 0.0122 (16) −0.0331 (17) C14 0.114 (2) 0.090 (2) 0.0513 (15) −0.0262 (17) −0.0139 (14) −0.0147 (14) C15 0.0842 (17) 0.0615 (13) 0.0469 (12) −0.0112 (12) −0.0106 (11) 0.0039 (10) C16 0.0449 (10) 0.0453 (10) 0.0433 (11) −0.0086 (8) 0.0050 (8) 0.0002 (8) C17 0.0689 (14) 0.0574 (13) 0.0476 (13) −0.0010 (11) 0.0028 (10) 0.0040 (9) C18 0.0793 (18) 0.096 (2) 0.0571 (15) −0.0071 (14) 0.0074 (13) 0.0312 (15) C19 0.0777 (19) 0.067 (2) 0.102 (3) −0.0175 (15) −0.0097 (18) 0.0453 (18) C20 0.0747 (17) 0.0393 (12) 0.117 (2) −0.0087 (11) −0.0142 (17) 0.0076 (13) C21 0.0597 (13) 0.0438 (11) 0.0685 (14) −0.0029 (9) −0.0012 (10) −0.0043 (9) C22 0.0402 (11) 0.0404 (10) 0.0469 (11) −0.0004 (7) −0.0049 (8) 0.0035 (8) C23 0.0565 (14) 0.0373 (11) 0.0431 (14) −0.0018 (7) 0.0031 (9) −0.0023 (7) C24 0.0648 (17) 0.087 (2) 0.0559 (17) −0.0105 (11) 0.0123 (13) −0.0073 (11) C25 0.096 (3) 0.100 (3) 0.058 (2) −0.0137 (14) 0.0288 (19) −0.0092 (12) C26 0.117 (3) 0.080 (2) 0.0407 (14) −0.0020 (14) 0.0075 (17) −0.0036 (9) C27 0.092 (3) 0.094 (2) 0.0523 (18) 0.0076 (14) −0.0194 (17) −0.0043 (11) C28 0.0617 (16) 0.0771 (18) 0.0475 (15) 0.0072 (11) −0.0006 (12) −0.0027 (9) N1 0.0623 (11) 0.0561 (10) 0.0750 (13) 0.0096 (9) 0.0040 (10) 0.0142 (9) N2 0.0497 (10) 0.0562 (10) 0.0737 (13) −0.0021 (8) −0.0049 (8) 0.0047 (8) O1 0.0475 (7) 0.0400 (6) 0.0559 (10) 0.0022 (6) −0.0022 (8) 0.0001 (9) Sn1 0.04407 (7) 0.03517 (6) 0.04012 (7) −0.00214 (4) 0.00003 (10) 0.00169 (12) Cl1 0.0463 (2) 0.0562 (2) 0.0904 (4) 0.00624 (17) 0.0004 (4) 0.0133 (5)
Geometric parameters (Å, º)
C1—C2 1.347 (4) C15—C16 1.388 (3)
C1—C5 1.372 (3) C15—H15 0.9300
C1—H1 0.9300 C16—Sn1 2.124 (2)
C2—C3 1.355 (4) C17—C18 1.382 (3)
C2—H2 0.9300 C17—C22 1.384 (3)
C3—C4 1.354 (4) C17—H17 0.9300
C3—H3 0.9300 C18—C19 1.357 (4)
C4—N1 1.312 (3) C18—H18 0.9300
supporting information
sup-6
Acta Cryst. (2006). E62, m172–m174
C5—N1 1.323 (3) C19—H19 0.9300
C5—H5 0.9300 C20—C21 1.386 (3)
C6—N2 1.326 (3) C20—H20 0.9300
C6—C7 1.358 (4) C21—C22 1.387 (3)
C6—H6 0.9300 C21—H21 0.9300
C7—C8 1.361 (4) C22—Sn1 2.127 (2)
C7—H7 0.9300 C23—C28 1.387 (4)
C8—C9 1.362 (4) C23—C24 1.390 (4)
C8—H8 0.9300 C23—Sn1 2.145 (3)
C9—C10 1.372 (3) C24—C25 1.387 (5)
C9—H9 0.9300 C24—H24 0.9300
C10—N2 1.318 (3) C25—C26 1.356 (6)
C10—H10 0.9300 C25—H25 0.9300
C11—C16 1.395 (3) C26—C27 1.357 (6)
C11—C12 1.398 (3) C26—H26 0.9300
C11—H11 0.9300 C27—C28 1.387 (5)
C12—C13 1.371 (4) C27—H27 0.9300
C12—H12 0.9300 C28—H28 0.9300
C13—C14 1.356 (4) O1—Sn1 2.3469 (14)
C13—H13 0.9300 O1—H1B 0.76 (3)
C14—C15 1.382 (3) O1—H1A 0.78 (3)
C14—H14 0.9300 Sn1—Cl1 2.5068 (9)
C2—C1—C5 118.7 (3) C22—C17—H17 119.6
C2—C1—H1 120.7 C19—C18—C17 120.8 (3)
C5—C1—H1 120.7 C19—C18—H18 119.6
C1—C2—C3 118.7 (2) C17—C18—H18 119.6
C1—C2—H2 120.6 C18—C19—C20 119.7 (3)
C3—C2—H2 120.6 C18—C19—H19 120.2
C4—C3—C2 119.1 (2) C20—C19—H19 120.2
C4—C3—H3 120.5 C19—C20—C21 120.1 (3)
C2—C3—H3 120.5 C19—C20—H20 119.9
N1—C4—C3 123.7 (3) C21—C20—H20 119.9
N1—C4—H4 118.1 C20—C21—C22 120.9 (2)
C3—C4—H4 118.1 C20—C21—H21 119.5
N1—C5—C1 123.1 (3) C22—C21—H21 119.5
N1—C5—H5 118.4 C17—C22—C21 117.67 (19)
C1—C5—H5 118.4 C17—C22—Sn1 120.99 (15)
N2—C6—C7 124.0 (2) C21—C22—Sn1 121.32 (16)
N2—C6—H6 118.0 C28—C23—C24 117.0 (3)
C7—C6—H6 118.0 C28—C23—Sn1 122.0 (2)
C6—C7—C8 118.3 (2) C24—C23—Sn1 121.0 (2)
C6—C7—H7 120.8 C25—C24—C23 120.7 (3)
C8—C7—H7 120.8 C25—C24—H24 119.6
C7—C8—C9 119.1 (3) C23—C24—H24 119.6
C7—C8—H8 120.4 C26—C25—C24 120.8 (3)
C9—C8—H8 120.4 C26—C25—H25 119.6
supporting information
sup-7
Acta Cryst. (2006). E62, m172–m174
C8—C9—H9 120.8 C25—C26—C27 119.8 (4)
C10—C9—H9 120.8 C25—C26—H26 120.1
N2—C10—C9 123.4 (3) C27—C26—H26 120.1
N2—C10—H10 118.3 C26—C27—C28 120.2 (4)
C9—C10—H10 118.3 C26—C27—H27 119.9
C16—C11—C12 120.3 (2) C28—C27—H27 119.9
C16—C11—H11 119.9 C27—C28—C23 121.4 (3)
C12—C11—H11 119.9 C27—C28—H28 119.3
C13—C12—C11 119.9 (3) C23—C28—H28 119.3
C13—C12—H12 120.1 C4—N1—C5 116.6 (2)
C11—C12—H12 120.1 C10—N2—C6 116.7 (2)
C14—C13—C12 120.6 (3) Sn1—O1—H1B 115.2 (18)
C14—C13—H13 119.7 Sn1—O1—H1A 118.7 (18)
C12—C13—H13 119.7 H1B—O1—H1A 116 (3)
C13—C14—C15 120.1 (3) C16—Sn1—C22 116.54 (9)
C13—C14—H14 119.9 C16—Sn1—C23 122.39 (7)
C15—C14—H14 119.9 C22—Sn1—C23 119.84 (7)
C14—C15—C16 121.3 (2) C16—Sn1—O1 85.41 (7)
C14—C15—H15 119.3 C22—Sn1—O1 83.32 (7)
C16—C15—H15 119.3 C23—Sn1—O1 90.01 (9)
C15—C16—C11 117.8 (2) C16—Sn1—Cl1 92.74 (6) C15—C16—Sn1 120.33 (16) C22—Sn1—Cl1 93.72 (5) C11—C16—Sn1 121.88 (16) C23—Sn1—Cl1 94.59 (7) C18—C17—C22 120.8 (2) O1—Sn1—Cl1 175.34 (8) C18—C17—H17 119.6
C5—C1—C2—C3 0.1 (5) C24—C23—C28—C27 0.6 (3) C1—C2—C3—C4 −0.1 (5) Sn1—C23—C28—C27 −179.56 (16)
C2—C3—C4—N1 −0.7 (5) C3—C4—N1—C5 1.3 (4)
C2—C1—C5—N1 0.7 (5) C1—C5—N1—C4 −1.3 (4)
N2—C6—C7—C8 −1.7 (4) C9—C10—N2—C6 −0.1 (4)
C6—C7—C8—C9 1.7 (4) C7—C6—N2—C10 0.9 (4)
supporting information
sup-8
Acta Cryst. (2006). E62, m172–m174
C20—C21—C22—C17 1.1 (3) C28—C23—Sn1—C16 −103.04 (16) C20—C21—C22—Sn1 179.48 (17) C24—C23—Sn1—C16 76.77 (17) C28—C23—C24—C25 0.5 (3) C28—C23—Sn1—C22 63.82 (17) Sn1—C23—C24—C25 −179.27 (17) C24—C23—Sn1—C22 −116.37 (16) C23—C24—C25—C26 −0.9 (4) C28—C23—Sn1—O1 −18.48 (15) C24—C25—C26—C27 0.1 (4) C24—C23—Sn1—O1 161.33 (16) C25—C26—C27—C28 1.1 (4) C28—C23—Sn1—Cl1 160.78 (14) C26—C27—C28—C23 −1.5 (3) C24—C23—Sn1—Cl1 −19.42 (15)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A