Copyright © 1998, American Society for Microbiology
Nonperinatal Nosocomial Transmission of Candida albicans in a
Neonatal Intensive Care Unit: Prospective Study
SUSAN E. REEF,1† BRENT A. LASKER,1* DONA S. BUTCHER,2MICHAEL M. MCNEIL,1
RUTH PRUITT,1HARRY KEYSERLING,3ANDWILLIAM R. JARVIS4
Division of Bacterial and Mycotic Diseases1and Hospital Infections Program,4Centers for Disease Control and
Prevention, Atlanta, Georgia 30333, and Divisions of Neonatology2and Pediatric Infectious Diseases and
Immunology,3Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia 30322
Received 15 September 1997/Returned for modification 28 November 1997/Accepted 27 January 1998
Nosocomial Candida albicans infections have become a major cause of morbidity and mortality in neonates in neonatal intensive care units (NICUs). To determine the possible modes of acquisition of C. albicans in hospitalized neonates, we conducted a prospective study at Grady Memorial Hospital, Atlanta, Ga. Clinical samples for fungal surveillance cultures were obtained at birth from infants (mouth, umbilicus, and groin) and their mothers (mouth and vagina) and were obtained from infants weekly until they were discharged. All infants were culture negative for C. albicans at birth. Six infants acquired C. albicans during their NICU stay. Thirty-four (53%) of 64 mothers were C. albicans positive (positive at the mouth, n526; positive at the vagina,
n518; positive at both sites, n510) at the time of the infant’s delivery. A total of 49 C. albicans isolates were analyzed by restriction endonuclease analysis and restriction fragment length polymorphism analysis by using genomic blots hybridized with the CARE-2 probe. Of the mothers positive for C. albicans, 3 of 10 were colonized with identical strains at two different body sites, whereas 7 of 10 harbored nonidentical strains at the two different body sites. Four of six infants who acquired C. albicans colonization in the NICU had C. albicans-positive mothers; specimens from all mother-infant pairs had different restriction endonuclease and CARE-2 hybridization profiles. One C. albicans-colonized infant developed candidemia; the colonizing and infecting strains had identical banding patterns. Our study indicates that nonperinatal nosocomial transmission of C.
albicans is the predominant mode of acquisition by neonates in NICUs at this hospital; mothers may be
colonized with multiple strains of C. albicans simultaneously; colonizing C. albicans strains can cause invasive disease in neonates; and molecular biology-based techniques are necessary to determine the epidemiologic relatedness of maternal and infant C. albicans isolates and to facilitate determination of the mode of transmission.
The rate of invasive candidiasis has increased substantially over the past decade (3). Patient populations identified to be at risk for candidemia include those who are immunocompro-mised because of disease, such as human immunodeficiency virus infection, or underlying conditions, such as malignancy, burns, hematologic disorders, or solid-organ and bone marrow transplantation. Also at risk are very low birth weight infants (birth weight, ,1,500 g) and infants cared for in neonatal intensive care units (NICUs). The most common manifesta-tions of Candida sp. infecmanifesta-tions in neonates are thrush and cutaneous lesions; however, candidemia in neonates is an in-creasingly important problem that may be associated with both morbidity and mortality (8, 29). Prevention of candidemia in neonates has been aided by the identification of specific risk factors (32). Some of these include the rate of colonization, which is higher in pregnant women than nonpregnant women (19), decreased T-cell, macrophage, and neutrophil function (4, 9, 33), use of peripheral venous catheters (15), and the presence of necrotizing enterocolitis (26). Prolonged hospital-ization, use of broad-spectrum antimicrobial agents, low birth weights, use of central, arterial, and venous catheters, and
prolonged parenteral nutrition have also been identified as risk factors for neonates (1, 2, 12).
While specific risk factors for candidemia in neonates have been identified, little is known about the epidemiology of can-didemia in NICUs. Numerous outbreaks of invasive Candida albicans infections in NICU patients have been described (5, 7, 10, 11, 21, 25, 30). Several of the phenotypic methods used to type outbreak-related isolates have lacked adequate discrimi-natory power or reliability compared with genotypic typing methods (13). Molecular biology-based typing of selected out-break-related isolates has implicated contaminated syringe flu-ids (25) or cross infection from the hands of health care work-ers (5, 7, 21) as the source of infection. Few prospective studies have been performed to identify the source and modes of transmission of C. albicans in NICU patients, and few studies have used molecular biology-based typing methods to deter-mine isolate relatedness.
It has been hypothesized that the acquisition of C. albicans by neonates occurs by two possible routes: by mother-neonate transmission or via environmental sources (cross contamina-tion). In this investigation, C. albicans isolates from mothers and neonates were collected prospectively and were then ex-amined by restriction enzyme analysis (REA) and hybridiza-tion with a digoxigenin-labeled CARE-2 probe to determine whether neonates acquired C. albicans perinatally from their mothers or postnatally from a nonmaternal source. Our data suggest that nonperinatal nosocomial transmission of C. albi-cans occurs in neonates.
(Part of this work was presented at the 91st General Meeting
* Corresponding author. Mailing address: Division of Bacterial and Mycotic Diseases, Building 1, Room 2225, Mailstop D-11, 1600 Clifton Rd., Centers for Disease Control and Prevention, Atlanta, GA 30333. Phone: (404) 639-2842. Fax: (404) 639-4421. E-mail: BAL3@CDC .GOV.
† Present address: National Immunization Program, Centers for Disease Control and Prevention, Atlanta, GA 30333.
1255
on May 15, 2020 by guest
http://jcm.asm.org/
of the American Society for Microbiology, Dallas, Texas, 5 to 9 May 1991 [22].)
MATERIALS AND METHODS
Epidemiologic surveillance.The investigation described here was approved by the Emory University School of Medicine Institutional Review Board. The study was conducted at Grady Memorial Hospital in Atlanta, Ga. Grady Memorial Hospital is a tertiary-care center in a county with a large population and has approximately 6,000 discharges annually. From 10 June to 23 June 1990, mother-term neonate pairs in which the neonate was delivered between 8:00 p.m. and 8:00 a.m. and all mother-preterm neonate pairs (24 h per day) were eligible for enrollment. Demographic information about the mothers and neonates was obtained by interviewing the mother and reviewing the infants’ medical records. Oral and vaginal samples for culture were obtained from the mother within 24 h after delivery. Within 24 h of admission, samples from the oropharynx, umbilicus, groin, and intravenous site of the neonate were obtained for culture by using a premoistened cotton-tipped swab. Subsequently, samples for culture were ob-tained from the same four sites of the neonates weekly until the time of discharge or death (the last infant enrolled in the study exited the study on 1 November 1990).
Yeast strains and identification.The clinical samples were immediately inoc-ulated onto Sabouraud dextrose agar (Difco Laboratories, Detroit, Mich.) and transported to the Centers for Disease Control and Prevention for the isolation of yeasts. The yeast isolates used in this investigation are listed in Table 1, along with their sources. Yeasts were identified by using the API 20C (bioMerieux Vitek, Inc., Hazelwood, Mo.) assimilation tests, germ tube induction in serum at 37°C, and microscopic examination of cells grown on cornmeal agar, as described by Buckley (6). Yeast stocks were maintained at 4°C on Sabouraud dextrose agar (Difco Laboratories).
Isolation of genomic DNA.All Candida sp. isolates were grown for 20 to 24 h at 28°C on a rotary shaker at 150 rpm in 10 ml of yeast peptone dextrose broth (14) in 50-ml flasks. The cells were harvested by centrifugation and washed once with sterile distilled water. Spheroplasts were then produced by the method of Scherer and Stevens (24). Spheroplasts were lysed and genomic DNA was pu-rified by repeated phenol-chloroform extractions as described previously by Mason et al. (18).
REA.DNA concentrations were estimated spectrophotometrically from the
A260/A280ratio and were digested with 10 U of EcoRI or ScaI (New England
Biolabs, Beverly, Mass.) permg for 6 h at 37°C in the buffer solution recom-mended by the manufacturer. DNA fragments were separated on 0.7% (wt/vol) agarose (International Biotechnologies Inc., New Haven, Conn.) gels in Tris-borate buffer (17). Following electrophoresis, the agarose gels were stained with ethidium bromide and were photographed with Polaroid type 55 film.
Ethidium bromide-stained EcoRI restriction fragments were compared by using enlarged prints (5 by 7 in.) made from the negative image. DNA from C.
albicans H317 was included on each gel to serve as a control standard. The
enlarged prints were examined visually for differences or similarities of bands between isolates as described by Reagan et al. (20). Following visual examina-tion, each unique EcoRI DNA type was given a number from 1 to 19.
CARE-2 analysis.Restriction fragments were transferred to nitrocellulose filters (type BA85; Schleicher & Schuell, Inc., Keene, N.H.) as described by Maniatis et al. (17). The CARE-2 DNA probe was labeled with digoxigenin as described in the Genius kit (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Conditions for hybridization, washing, signal detection, and photography have been described previously (14). The restriction fragment length polymor-phism (RFLP) patterns obtained with the CARE-2 probe (CARE-2 pattern) were detected by visual inspection of the filters. Then, each CARE-2 pattern was given a letter from A to AA.
RESULTS
Epidemiologic study. A total of 64 mother-neonate pairs were enrolled in the study. This included 57 term neonates (mean gestational age, 39 weeks; mean birth weight, 3,078 g) and 7 preterm neonates (mean gestational age, 29 weeks; mean birth weight, 1,093 g). No samples (0 of 256) from the infants were culture positive for C. albicans within 2 days of delivery. Samples from 34 of 64 (53%) mothers (a total of 50 of 128 [39%] samples from the mothers) were culture positive for C. albicans within 2 days of delivery. Twenty-six mothers were colonized orally, 18 were colonized vaginally, and 10 were colonized at both sites. Among the infants, a total of 30 of 256 samples were positive for fungi: 15 of 30 positive samples from six infants were positive for C. albicans. Four of these six infants had mothers who were culture positive for C. albicans within 2 days of delivery. The remaining 15 positive samples,
obtained from eight different infants, were positive for either Candida parapsilsosis, Candida lusitaniae, or Candida glabrata; these isolates were not characterized further.
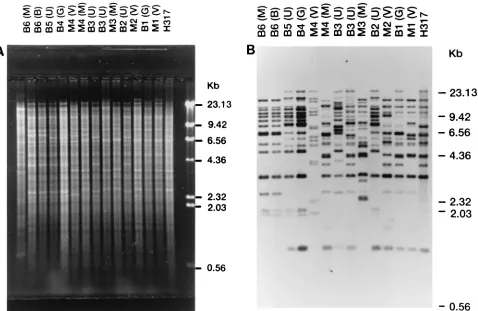
REA analysis.A total of 17 different EcoRI DNA types were found among the 46 C. albicans isolates (22 from mothers, 23 from neonates, and 1 reference strain [strain H317]). Different DNA types were observed among isolates from three of four mother-infant pairs. A single mother-infant pair (M1-B1) were colonized with isolates with identical EcoRI DNA types (Fig. 1A). The EcoRI DNA types of serial C. albicans isolates
ob-TABLE 1. Distribution of genotypes of C. albicans isolates from mothers and neonates by site
Isolate no. Patient (sitea) EcoRI pattern CARE-2 pattern
H317 17 I
1 M1 (M) 1 J
2 M1 (V) 1 K
3 B1 (U1) 1 L
4 B1 (G1) 1 L
5 B1 (U2) 1 L
6 B1 (G2) 1 L
7 M2 (V) 1 M
8 B2 (U1) 2 N
9 B2 (G1) 2 N
10 B2 (M) 2 N
11 B2 (U2) 2 N
12 B2 (G2) 2 N
13 M3 (M) 3 O
14 B3 (U1) 1 P
15 B3 (G1) 1 P
16 B3 (M) 1 P
17 B3 (U2) 4 Q
18 B3 (G2) 1 R
19 M4 (M) 1 S
20 M4 (V) 5 T
21 B4 (U1) 6 Cpb
22 B4 (G1) 6 Cp
23 B4 (U2) 6 Cp
24 B4 (G2) 7 U
25 B5 (U) 7 U
26 B5 (G) 7 U
27 B6 (M1) 8 V
28 B6 (U1) 8 V
29 B6 (G1) 9 Tgc
30 B6 (M2) 8 V
31 B6 (U2) 8 V
32 B6 (G2) 8 V
33 B6 (B) 8 V
34 M7 (M) 18 X
35 M7 (V) 10 Y
36 M8 (M) 11 Z
37 M8 (V) 12 AA
38 M9 (M) 1 A
39 M9 (V) 1 W
40 M10 (M) 13 B
41 M10 (V) 13 B
42 M11 (M) 14 C
43 M11 (V) 14 C
44 M12 (M) 1 D
45 M12 (V) 19 E
46 M13 (M) 15 F
47 M13 (V) 15 G
48 M14 (M) 16 H
49 M14 (V) 16 H
aB, blood; G, groin; M, mouth; U, umbilicus; V, vagina. Numbers in
paren-theses for a given site represent serial isolates.
bCp, C. parapsilosis. cTg, T. glabrata.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:2.612.309.546.91.557.2]tained from serial samples from a given site from a patient were generally persistent; however, a different EcoRI type was observed for a later isolate obtained from the umbilicus from baby 3 (isolate U1 versus isolate U2 in baby B3). Two neonates (babies B4 and B6) acquired mixed yeast infections. Baby B4 was colonized with C. albicans and C. parapsilosis, whereas baby B6 was colonized with C. albicans and C. glabrata. Baby B6 became colonized with C. albicans after birth and subse-quently developed candidemia. Invasive candidiasis was docu-mented at autopsy. The C. albicans isolates colonizing baby B6 and infecting that baby’s bloodstream were identical (i.e., EcoRI DNA type 8).
When we analyzed by their EcoRI patterns the isolates ob-tained from 10 mothers from whom clinical samples from two different body sites, i.e., the vagina and the mouth, were cul-ture positive, 6 mothers were found to be colonized with iden-tical C. albicans strains and 4 mothers were found to harbor nonidentical C. albicans strains at the two different body sites (Table 1).
Analysis with the CARE-2 probe.The degree of relatedness among our panel of C. albicans isolates was also determined by analyzing the RFLP patterns obtained with the CARE-2 probe (CARE-2 patterns) since this C. albicans-specific multiple-gene-family probe was previously shown to be highly polymor-phic (14, 16). A total of 27 different CARE-2 patterns were observed, and these are listed in Table 1. Surprisingly, the isolates from EcoRI-digested samples from all four mother-infant pairs had nonidentical CARE-2 patterns (Fig. 1B). The
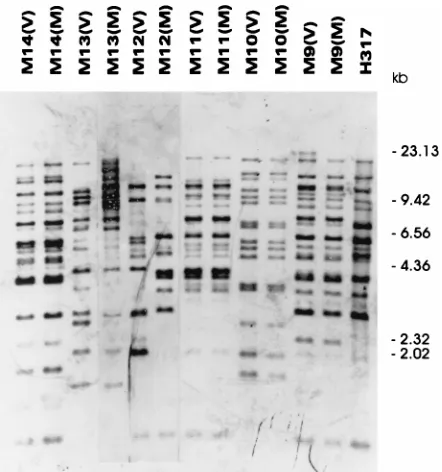
nonidentical CARE-2 patterns between isolates from the mothers and the infants were confirmed by hybridization of the CARE-2 probe to ScaI-digested samples (Fig. 2). Interestingly, the isolates from babies B4 and B5, who were in the NICU at the same time, were observed to have identical CARE-2 pat-terns (Fig. 1B and 2). The CARE-2 banding patpat-terns of iso-lates obtained from the blood and several colonizing sites from baby B6, who developed invasive candidiasis and died, were identical (Fig. 1B and 2). Analysis of the CARE-2 patterns for isolates from 10 mothers who were colonized at two different body sites showed that for 7 mothers the isolates from the different sites had nonidentical CARE-2 patterns (Fig. 3 and Table 1).
DISCUSSION
[image:3.612.62.540.68.379.2]Despite an increase in systemic Candida sp. infections ob-served in neonates, few prospective studies have evaluated by molecular biology-based epidemiologic methods either the source or the mode of transmission. By using two different typing methods, we demonstrated that all maternal and neo-natal colonizing strains were nonidentical; thus, colonization of neonates with C. albicans occurred primarily after delivery in this particular NICU. Nonperinatal nosocomial transmission of C. albicans suggests that the route of transmission is primar-ily from nonmaternal sources, possibly via cross contamination of the hands of health care workers or parents. Determination
FIG. 1. REA and RFLP analysis with the CARE-2 probe. (A) Ethidium bromide-stained gel of EcoRI-digested genomic DNA from strains from mother-baby pairs. (B) DNA fragments from the gel in panel A were transferred to a sheet of nitrocellulose and then hybridized with the CARE-2 probe. Lanes are labeled M1 to M4 for each of four mothers, respectively, and B1 to B6 for each of six neonates, respectively; the symbols in parentheses denote sample sites: B, blood; M, mouth; G, groin; U, umbilicus; and V, vagina. Bacteriophage lambda DNA digested with HindIII was used as a molecular size markers, and the molecular sizes are indicated on the right.
on May 15, 2020 by guest
http://jcm.asm.org/
of the source of exogenous infection and mode of transmission requires further investigation.
This study reemphasizes why it is important for health care workers to wash their hands and follow other infection control procedures to prevent the nosocomial transmission of patho-gens in the NICU environment. An endogenous source of candidemia has been suggested in an outbreak of candidemia in an NICU (20). Our findings differ from those of Waggoner-Fountain et al. (31), who observed the same DNA types among three of four isolates from mothers and infants by REA of genomic DNA and electrophoretic karyotype analysis. The differences in the mode of transmission observed between these two groups of investigators may be due in part to differ-ences in typing methods, the sites from which samples were obtained, patient enrollment, patient demographics, infection control practices, and sample size. These differences demon-strate two different modes of acquisition of yeast colonization in the NICU: maternal transmission (vertical transmission) and nonperinatal nosocomial transmission (horizontal trans-mission). In our study, an environmental source of transmis-sion in the NICU was also suggested, since the organisms colonizing babies B4 and B5 (C. parapsilosis and T. glabrata, respectively) were not consistent with the species colonizing the respective mothers. This is consistent with a previous out-break of C. parapsilosis in an NICU caused by cross infection (23).
Another important observation from this investigation con-cerns different strains isolated from different body sites from 7 of the 10 mothers analyzed. Different strains at different body locations of the same person were observed previously (27, 28), indicating a more complex pattern of carriage than was origi-nally hypothesized. This finding further complicates the assess-ment of sources and modes of transmission and indicates that samples from multiple sites may need to be cultured and that multiple strains from one person may need to be genotyped.
One neonate colonized with a C. albicans isolate after birth developed candidemia, and invasive candidiasis was docu-mented at autopsy. Examination of the EcoRI and CARE-2
patterns of the isolate from this infant demonstrated identical patterns for the colonizing and bloodstream isolates, indicating that the colonizing strain was the source of the invasive disease. For the majority of patients, we observed that serial isolates had identical DNA types according to their CARE-2 patterns. However, for one infant, baby B3, we found that two different strains colonized the umbilicus. Strains with different DNA types at the same site may be due either to strain replacement or to the coexistence of multiple strains at the site of infection. Interestingly, two infants, babies B4 and B5, were colonized with the same strain, as demonstrated by identical EcoRI and CARE-2 patterns, yet the colonizing strains differed from the strains from their mothers. Since both babies occupied the same NICU quadrant at the same time, this is further evidence supporting the horizontal transmission of C. albicans in this NICU. Nosocomial transmission of Candida by cross contam-ination or via common contaminated sources has been re-ported previously (5, 7, 21, 25).
Analysis of the CARE-2 patterns better discriminated among isolates of C. albicans than REA with EcoRI. For in-stance, we identified 27 different CARE-2 patterns but only 17 different patterns by REA with EcoRI. The higher degree of discrimination is due to the ability of CARE-2 to discriminate among isolates with the same EcoRI DNA types. A high de-gree of discrimination of strains of C. albicans according to the CARE-2 patterns was also observed by Lockhart et al. (16).
Our data indicate that postnatal acquisition of C. albicans by neonates more commonly occurs by transmission in NICUs than by transmission from mothers to their infants. Thus, rather than focusing on methods of identifying pregnant women with vaginal C. albicans colonization and instituting prevention interventions to interrupt the transmission to their infants during delivery, our infection control interventions should focus on high-risk neonates and on hand washing by health care workers.
In summary, our data demonstrate that (i) the predominant mode of acquisition of C. albicans in the NICU is by
[image:4.612.55.283.69.266.2]nonperi-FIG. 2. Hybridization of ScaI-digested genomic DNA with the CARE-2 probe. Lanes are labeled M1 to M4 for each of four mothers, respectively, and B1 to B6 for each of six neonates, respectively; the symbols in parentheses denote sample sites: B, blood; M, mouth; G, groin; U, umbilicus; and V, vagina. Bac-teriophage lambda DNA digested with HindIII was used as a molecular size marker, and the molecular sizes are indicated on the right.
FIG. 3. Hybridization of EcoRI-digested genomic DNA with the CARE-2 probe. Lanes are labeled M9 to M14 for each of six different mothers, respec-tively; the symbols in parentheses denote sample sites: M, mouth, and V, vagina.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:4.612.318.539.461.697.2]natal nosocomial transmission rather than by mother-infant transmission, (ii) the colonizing strain may be the source of invasive disease, and (iii) mothers and babies may be colonized with more than one strain at different body sites. These data demonstrate the utility of molecular biology-based typing methods for enhancing our understanding of the epidemiology of nosocomial C. albicans infections. These methods, including the application of REA and RFLP analysis with the CARE-2 gene probe, will undoubtedly be useful for further defining risk factors for invasive diseases, identifying potentially virulent strains, and developing and assessing interventions that will prevent transmission.
REFERENCES
1. Baley, J. E., R. M. Kliegman, B. Boxerbaum, and A. A. Fanaroff. 1986. Fungal colonization in the very-low-birth-weight infant. Pediatrics 78:225– 232.
2. Baley, J. E., R. M. Kliegman, and A. A. Fanaroff. 1984. Disseminated fungal infections in low-birth-weight infants: clinical manifestations and epidemiol-ogy. Pediatrics 73:144–152.
3. Beck-Sague, C., W. R. Jarvis, and the National Nosocomial Infections
Sur-veillance System.1993. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. J. Infect. Dis. 167:1247– 1251.
4. Bektas, S., B. Goetze, and C. P. Speer. 1990. Decreased adherence, chemo-taxis and phagocytic activities of neutrophils from preterm neonates. Acta Pediatr. Scand. 79:1031–1038.
5. Betremieux, P., S. Chevrier, G. Quindos, D. Sullivan, L. Polonelli, and C.
Guiguen.1994. Use of DNA fingerprinting and biotyping methods to study a Candida albicans outbreak in a neonatal intensive care unit. Pediatr. Infect. Dis. J. 13:899–905.
6. Buckley, H. R. 1989. Identification of yeast, p. 97–109. In E. G. V. Evans and M. D. Richardson (ed.), Medical mycology: a practical approach. IRL Press at Oxford University Press, Oxford, England.
7. Burnie, J. P., F. C. Odds, W. Lee, C. Webster, and J. D. Williams. 1985. Outbreak of systemic Candida albicans in intensive care unit caused by cross infection. Br. Med. J. 290:746–748.
8. Butler, K. M., and C. J. Baker. 1988. Candida: an increasingly important pathogen in the nursery. Pediatr. Clin. N. Am. 35:543–563.
9. D’Ambola, J. B., M. P. Sherman, D. P. Tashkin, and H. Gong. 1988. Human and rabbit newborn lung macrophages have reduced anti-Candida activity. Pediatr. Res. 24:285–290.
10. Faix, R. G., D. J. Finkel, R. D. Andersen, and M. K. Hostetter. 1995. Genotypic analysis of a cluster of systemic Candida albicans infections in a neonatal intensive care unit. Pediatr. Infect. Dis. J. 14:1063–1068. 11. Fraser, C. A. M. 1990. Cross-infection and diversity of Candida albicans
strain carriage in patients and nursing staff on an intensive care unit. J. Vet. Med. Mycol. 28:317–325.
12. Herruzo-Cabrera, R., C. De-Lope, M. Ferna´ndez-Arjona, and J. Rey-Calero. 1995. Risk factors of infection and digestive tract colonization by Candida spp. in a neonatal intensive care unit. Eur. J. Epidemiol. 11:291–295. 13. Hunter, P. R. 1991. A critical review of typing methods for Candida albicans
and their applications. Crit. Rev. Microbiol. 17:417–434.
14. Lasker, B. A., L. S. Page, T. J. Lott, and G. S. Kobayashi. 1992. Isolation, characterization, and sequencing of Candida albicans repetitive element 2. Gene 116:51–57.
15. Leibovitz, E., A. Luster-Reicher, M. Amitai, and B. Mogilner. 1992. Systemic candidal infections associated with use of peripheral venous catheters in
neonates: a 9-year experience. Clin. Infect. Dis. 14:485–491.
16. Lockhart, S. R., B. D. Reed, C. L. Pierson, and D. R. Soll. 1996. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE-2. J. Clin. Microbiol. 34:767–777.
17. Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
18. Mason, M. M., B. A. Lasker, and W. S. Riggsby. 1987. Molecular probe for the identification of medically important Candida species and Torulopsis
glabrata. J. Clin. Microbiol. 25:563–566.
19. Miller, M. J. 1990. Fungal infections, p. 475–515. In J. S. Remington and J. O. Klein (ed.), Infectious disease of the fetus and newborn, 3rd ed. The W. B. Sanders Co., Philadelphia, Pa.
20. Reagan, D. R., M. A. Pfaller, R. J. Hollis, and R. P. Wenzel. 1990. Charac-terization of the sequence of colonization and nosocomial candidemia using DNA fingerprinting and a DNA probe. J. Clin. Microbiol. 28:2733–2738. 21. Reagan, D. R., M. A. Pfaller, R. J. Hollis, and R. P. Wenzel. 1995. Evidence
of nosocomial spread of Candida albicans causing bloodstream infection in a neonatal intensive care unit. Diagn. Microbiol. Infect. Dis. 21:191–194. 22. Reef, S., S. Butcher, B. Lasker, M. McNeil, R. Pruit, H. Keyserling, and W.
Jarvis.1991. Investigation of the role of perinatal transmission of Candida
albicans (CA) in neonates by restriction endonuclease analysis (REA) and a
digoxigenin-labeled CA repetitive element (CARE-2) probe, abstr. L-27, p. 427. In Program and abstracts of the 91st General Meeting of the American Society for Microbiology 1991. American Society for Microbiology, Wash-ington, D.C.
23. Saxen, H., M. Virtanen, P. Carlson, K. Hoppu, M. Pohjavuori, M. Vaara, J.
Vuopio-Varkila, and H. Peltola.1995. Neonatal Candida parapsilosis out-break with a high case fatality rate. Pediatr. Infect. Dis. J. 14:776–781. 24. Scherer, S., and D. A. Stevens. 1988. A Candida albicans dispersed, repeated
gene family and its epidemiologic application. Proc. Natl. Acad. Sci. USA
85:1452–1456.
25. Sherertz, R. J., K. S. Gledhill, K. D. Hampton, M. A. Pfaller, L. B. Givner,
J. S. Abramson, and R. G. Dillard.1992. Outbreak of Candida bloodstream infections associated with retrograde medication administration in a neona-tal intensive care unit. J. Pediatr. 120:455–461.
26. Smith, S. D., E. P. Tagge, J. Miller, H. Cheu, K. Sukarochana, and M. I.
Rowe.1990. The hidden mortality in surgically treated necrotizing entero-colitis: fungal sepsis. J. Pediatr. Surg. 25:1030–1033.
27. Soll, D. R., C. J. Langtimm, J. McDowell, J. Hicks, and R. Galask. 1987. High-frequency switching in Candida strains isolated from vaginitis patients. J. Clin. Microbiol. 25:1611–1622.
28. Soll, D. R., M. Staebell, C. Langtimm, M. Pfaller, J. Hicks, and G. Roa. 1988. Multiple Candida strains in the course of a single systemic infection. J. Clin. Microbiol. 26:1448–1459.
29. Stamos, J. K., and A. H. Rowley. 1995. Candidemia in a pediatric population. Clin. Infect. Dis. 20:571–575.
30. Vaudry, W. L., A. J. Tierney, and W. M. Wenman. 1988. Investigation of a cluster of systemic Candida albicans infections in a neonatal intensive care unit. J. Infect. Dis. 158:1375–1379.
31. Waggoner-Fountain, L. A., M. W. Walker, R. J. Hollis, M. A. Pfaller, J. E.
Ferguson II, R. P. Wenzel, and L. G. Donowitz.1996. Vertical and horizontal transmission of unique Candida species to premature newborns. Clin. Infect. Dis. 22:803–808.
32. Weese-Mayer, D. E., D. W. Fondriest, R. T. Brouillette, and S. T. Shulman. 1987. Risk factors associated with candidemia in the neonatal intensive care unit: a case-control study. Pediatr. Infect. Dis. 6:190–196.
33. Wilson, C. B. 1986. Immunologic basis for increased susceptibility of the neonate to infection. J. Pediatr. 108:1–12.