This may be the author’s version of a work that was submitted/accepted for publication in the following source:
Sutherland, Heidi, Maher, Bridget, Rodriguez Acevedo, Astrid Jannet,
Haupt, Larisa, &Griffiths, Lyn
(2014)
Investigation of brain-derived neurotrophic factor (BDNF) gene variants in migraine.
Headache,54(7), pp. 1184-1193.
This file was downloaded from: https://eprints.qut.edu.au/105616/
c
Consult author(s) regarding copyright matters
This work is covered by copyright. Unless the document is being made available under a Creative Commons Licence, you must assume that re-use is limited to personal use and that permission from the copyright owner must be obtained for all other uses. If the docu-ment is available under a Creative Commons License (or other specified license) then refer to the Licence for details of permitted re-use. It is a condition of access that users recog-nise and abide by the legal requirements associated with these rights. If you believe that this work infringes copyright please provide details by email to qut.copyright@qut.edu.au
Notice:Please note that this document may not be the Version of Record (i.e. published version) of the work. Author manuscript versions (as Sub-mitted for peer review or as Accepted for publication after peer review) can be identified by an absence of publisher branding and/or typeset appear-ance. If there is any doubt, please refer to the published source.
1
Investigation of brain derived neurotrophic factor (BDNF) gene variants in migraine.
Heidi G. Sutherland, PhD; Bridget H. Maher, PhD; Astrid J. Rodriguez-Acevedo, BBiol; Larisa M.
Haupt, PhD; and Lyn R. Griffiths*, PhD
Genomics Research Centre, Institute of Health and Biomedical Innovation, Queensland University of
Technology, Musk Ave, Kelvin Grove, QLD 4059, Australia
* Corresponding author
Phone: +617 3138 6102
Fax: +617 3138 6039
Email: lyn.griffiths@qut.edu.au
Conflict of Interest Statement: The authors declare that there is no conflict of interest.
Keywords: BDNF, migraine, SNP, association study
Abbreviations: IHS – International Headache Society, MA – migraine with aura, MO – migraine
without aura, CSD – cortical spreading depression, BDNF – brain derived neurotrophic factor, CNS –
central nervous system, TGN – trigeminal ganglion neuron, CGRP- – calcitonin gene-related
polypeptide alpha, SNP – single nucleotide polymorphism, RFLP – restriction-fragment length
polymorphism, PCR – polymerase chain reaction, dNTPs – deoxynucleotide triphosphates, χ2 –
2 ABSTRACT
Objective. – A number of observations have suggested that brain derived neurotrophic factor (BDNF)
plays a role in migraine pathophysiology. This study investigates whether variants in the BDNF gene
are associated with migraine in an Australian case-control population.
Background. – Brain derived neurotrophic factor (BDNF) has an important role in neural growth,
development and survival in the central nervous system and is an important modulator of central and
peripheral pain responses. Variants in BDNF, in particular the functional Val66Met polymorphism
(rs6265), have been found to be associated with a number of psychiatric disorders, cognitive function
and obesity. As BDNF has been found to be differentially expressed in a number of aspects related to
migraine, we tested for association between single nucleotide polymorphisms (SNPs) in BDNF and
migraine.
Methods. – Five SNPs in the BDNF locus (rs1519480, rs6265, rs712507, rs2049046 and rs12273363)
were genotyped initially in a cohort of 277 migraine cases, including 172 diagnosed with migraine with
aura (MA) and 105 with migraine without aura (MO), and 277 age- and sex-matched controls. Three
of these SNPs (rs6265, rs2049046 and rs12273363) were subsequently genotyped in a second cohort
of 580 migraineurs, including 473 diagnosed with MA and 105 with MO, and 580 matched controls.
Results. – BDNF SNPs rs1519480, rs6265, rs712507 and rs12273363 were not significantly associated
with migraine. However, rs2049046 showed a significant association with migraine, and in particular,
MA in the first cohort. In the second cohort, although an increase in the rs2049046 T-allele frequency
was observed in migraine cases, and in both MA and MO subgroups, it was not significantly different
from controls. Analysis of data combined from both cohorts for rs2049046 showed significant
differences in the genotypic and allelic distributions for this marker in both migraine and the MA
sub-group.
Conclusion. – This study confirmed previous studies that the functional BDNF SNP rs6265 (Val66Met)
3
one the BDNF transcripts, may be associated with migraine, suggesting that further investigations of
4 INTRODUCTION
Migraine is a common episodic neurological disorder characterized by severe head pain, usually
accompanied by nausea, vomiting, neurological disturbance, photophobia and phonophobia.
Migraine affects approximately 6% of men and 17% of women and has significant personal, social and
economic burdens [1]. Two main types of migraine have been classified by the International Headache
Society [IHS], migraine with aura (MA) or without aura (MO) [2]. MA is distinguished by the presence
of neurological disturbances preceding headache in the early stages of the attack. These are usually
visual, taking the form of scintillating shapes, hallucinations or black spots. The pathophysiology of
migraine is only partially understood, but is believed to be caused by activation of the
trigeminovascular system which results in vasodilation of pain-producing intracranial blood vessels
and activation of second-order sensory neurons in the trigeminal nucleus caudalis with subsequent
perception of pain [3].
Brain derived neurotrophic factor (BDNF) has an important role in neuronal growth, developmental
and survival in the central nervous system (CNS) [4], and has also emerged as an important modulator
of central and peripheral nociceptor responses [5, 6]. The BDNF gene is complex; it contains 11 exons
that are alternatively spliced to encode many transcripts which are differentially expressed in diverse
tissues, and in various regions within the brain, including high expression in the CNS, including the
trigeminal ganglion neurons (TGNs) [7]. BDNF is synthesized as a precursor protein (proBDNF) that is
proteolytically cleaved to produce mature BDNF [8]. ProBDNF preferentially binds the receptor
p75NTR which can trigger apoptosis, axonal retraction and pruning dendritic spines and mature BDNF
binds the TrkB receptor tyrosine kinase which affects cell cycle, neurite outgrowth and synaptic
plasticity [9]. Decreased BDNF in serum is one of the best known biological markers of depression and
5
A number of observations have suggested that BDNF may also play a role in migraine. Studies have
shown that serum levels of BDNF are elevated both during a migraine attack [12], and when compared
to non-migraineur controls [13]. Migraineurs have also been found to have a significant decrease in
levels of BDNF in platelets compared to controls [14], possibly attributable to platelet activation during
migraine attacks with immediate release of BDNF [13]. BDNF is a mediator of activity-dependent
plastic changes in second-order trigeminal neurons that characterise the central sensitization
observed in migraine [15]. In rats BDNF is also co-expressed in TGNs with calcitonin gene-related
polypeptide alpha (CGRP-), a key vasodilating neuropeptide implicated in migraine; CGRP-
enhances the release of BDNF from TGNs, which can be abolished by a CGRP antagonist [16]. Finally,
BDNF was one of the genes found to be differentially expressed after cortical spreading depression
(CSD) in the rat cortex [17]. Cortical spreading depression is a slowly propagating wave of neuronal
and glial depolarisation which can activate trigeminal nociceptors, and is thought to underlie the aura
component of MA [18, 19].
.
Variants in BDNF have been implicated in the susceptibility to memory and hippocampal function
impairments [20, 21], and a number of psychiatric disorders including obsessive-compulsive disorder,
eating disorders, bipolar disorder, schizophrenia, major depression, and Alzheimer disease [22-24].
BDNF mutations and DNA variants have also been implicated in obesity and body mass index [25-28].
The most extensively studied variant is rs6265, the G196A single nucleotide polymorphism (SNP)
which is in the proprotein region of BDNF resulting in the substitution of a valine for a methionine
residue at position 66 (Val66Met). The polymorphism modifies the intracellular packaging of proBDNF
and impacts on activity-dependent secretion of mature BDNF [29]. To date two studies have
investigated whether SNPs at the BDNF locus are associated with migraine. In a case-control study on
a German cohort, no association was found for the rs6265 SNP with migraine overall, or either the MA
6
BDNF locus in a Spanish population; while they found no significant main effects they did detect an
increased risk between the AT-genotype of rs2049046 in BDNF and the GC-genotype of rs1553005 in
the CGRP gene, suggesting an interaction between these genes [31].
Because of the relationship between BDNF and CSD we hypothesised that polymorphisms in this gene
may increase risk of migraine, particularly the MA subtype. Therefore in this study we investigated a
range of SNPs in BDNF in an Australian migraine case-control population recruited from South-East
Queensland which has a particularly large component of MA cases.
7 Subjects
Two independent cohorts were used in this study. Cohort 1 consists of 277 migraineurs and 277
controls matched for sex, age (+/-5 years) and ethnicity, recruited from in and around South Eastern
Australia was used for this study [32]. Cohort 2 consists of 580 migraineurs and 580 sex-, age and
ethnicity-matched controls from South-Eastern Australia. All individuals provided informed consent
and were adult Caucasians of European decent living in Australia, having ancestors who emigrated
within the last 160 years from various locations within the British Isles and other parts of Europe.
Migraineurs were diagnosed by a clinical neurologist as having either MA or MO based strictly on the
widely accepted criteria specified by the IHS [2]. The study was approved by the Griffith University,
and subsequently the Queensland University of Technology, Ethics Committees for Experimentation
on Human Subjects. A whole blood sample was collected from each participant and genomic DNA was
extracted from white blood cells using a standard salting out method [33]. Samples used for the
genotyping study were all from unrelated individuals and the control group consisted of individuals
with no family history of migraine.
Genetic Analysis
Genotypes for rs6265 and rs2049046 were determined by restriction-fragment length polymorphism
(RFLP) analysis of restriction enzyme-digested PCR products on 3% agarose gels. A 20-l PCR reaction
mix contained 1xPCR buffer, 1.75 mM MgCl2, 0.2 mM dNTPs, 0.15 M of each primer, 20-40 ng of
genomic DNA and 1.5U of GoTaq® (Promega). Thermocycler conditions were an initial denaturation
at 95°C for 10 min, followed by 35 cycles of 95°C for 45 s, 60°C for 45s, 72°C for 45 s and a final
extension step of 72°C for 7 min. Specific primers were used to amplify each region containing the
SNPs and restriction enzymes (NEB) were chosen that would differentiate the two alleles after
8
rs6265 was genotyped as described previously [34] using the primers 5’-
CCTACAGTTCCACCAGGTGAGAAGAGT and 5’- GCTGCCGTTACCCACTCACT to generate a 480-bp PCR
product which with AflIII is digested into four fragments of 220-, 93-, 90- and 70-bp in the case of the
wild type “G” allele and only three fragments of 293-, 93- and 90-bp in the case of the “A” mutant
allele.
rs2049046 was genotyped using the primers 5’- ACCAATTTGTGCAGACCTTAAAA and 5’-
TCAAACCCCTTTGAGATTACAGA to generate a 353-bp PCR product which with HinfI is digested into
three fragments of 206-, 82- and 65-bp in the case of the wild type “T” allele and only two fragments
of 271- and 82-bp in the case of the mutant “A” allele.
Genotypes for rs1519480, rs7127507 and rs12273363 were genotyped on the Sequenom
MassARRAY® platform with the following primers:
5’ ACGTTGGATGCTGAAGAGTAAGAACAGATGC and 5’ ACGTTGGATGCTTAGGGAAATAAATGGAAGG and
extension primer 5’ TTTTTTCCTTAATGGCCC for rs1519480
5’ ACGTTGGATGTTAAAACATTCAAGCTTCC and 5’ ACGTTGGATGGAGAGAATAGAGAGTTGCGG and
extension primer 5’ TTCAAGCTTCCTTTCTACAA for rs7127507
5’ ACGTTGGATGGCTATTGACTGCAGGGATGA and 5’ ACGTTGGATGGCTGGGTGGTCTGAAACTTA and
extension primer 5’ CGATGCTGCAGAAGA for rs12273363
Statistical Analysis
Hardy-Weinberg equilibrium was verified for observed genotype frequencies for each SNP to detect
deviation from the normal genotype distribution in the population. Chi-square (χ2) analysis was
performed to test for significant differences in genotype and allele frequencies for each SNP in
migraineurs, MA and MO subgroups versus controls to detect any association with migraine. The
Statistical Package for Social Sciences (SPSS version 21.0) was used for statistical analyses. Haploview
9 RESULTS
BDNF is a complex locus on chromosome 11 which generates at least 17 alternatively spliced
transcripts and multiple alternatively spliced antisense transcripts (BDNF-AS) from the opposite
direction [7]. Initially five SNPs, covering 39 kb of the BDNF locus were genotyped in a migraine
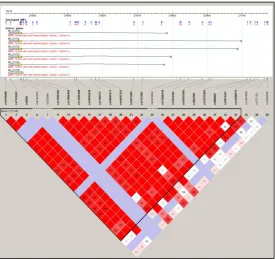
case-control population. Figure 1A shows the location of the SNPs with respect to a subset of BDNF
transcripts and the haploblock structure of the locus determined and visualised from HapMap data
using the genetic analysis program Haploview. In this study cohort 1 included 277 migraineurs, of
which 172 were diagnosed with MA and 105 with MO, and 277 matched controls. All five SNPs that
were genotyped were found to be in Hardy-Weinberg (p>0.05, data not shown). Our data is similar to
the Hapmap data which shows high linkage disequilibrium (LD) throughout the locus, with SNPs
covering the gene in a single block, although there is slightly different block definition for SNPs 5’ and
3’ to the locus (Figure 1B).
In Cohort 1 four of the SNPs (rs1519480, rs6265, rs7127507 and rs12273363) did not show any
association with migraine. When subgroup analysis was performed for these SNPs no associations
were detected with either MO or MA, although for rs6265 the numbers of individuals with the AA
genotype were too few (i.e. <5) in the MO or MA subgroups to perform chi-squared analysis and
similarly for the rs12273363 CC genotype in the MO group (Table 1). However, for rs2049046 some
significant differences in frequencies were detected between controls and migraineurs. For total
migraine, although the percentage of individuals with each genotype differed somewhat from the
control group, the p-value obtained from chi-squared analysis was not significant. However, when the
allele frequencies were compared there was a significant difference between control and migraine
groups with an increase in frequency of the minor T-allele in migraineurs (p=0.035). Subgroup analysis
10
individuals compared to controls, this was not significant. However, for MA, significant differences in
both genotype and allele frequencies were observed compared to controls (p=0.036 and p=0.0125,
respectively).
Cohort 2 was a migraine case-control population consisting of 580 migraineurs, including 107
individuals diagnosed with MO and 473 diagnosed with MA and 580 matched controls. This cohort
was used to investigate whether the positive association of migraine and, in particular, MA with the
T-allele of the BDNF rs2049046 SNP could be replicated in an independent, and larger, population.
BDNF rs6265 and rs12273363 were also genotyped in cohort 2 as they have low minor allele
frequencies, which meant that a valid chi-squared analysis was not able to be performed in Cohort 1
for some of the subgroups. In cohort 2, genotypic and allelic frequencies for rs6265 and rs12273363
were not significantly different between controls and individuals with migraine or those that suffer
from MA. Similarly for those individuals with MO, no association was found with the rs6265 A-allele.
For the BDNF rs2049046, an increase in the frequency of the TT genotype and T-allele in migraineurs
was detected compared to controls, similar to what was observed in Cohort 1. However, the increase
was smaller and was not significant in either total migraine, or the MA and MO subgroups. Hence the
interesting results from cohort 1 for this marker were not replicated in cohort 2.
Analysis of the data from Cohorts 1 and 2 (Table 3) together also supports the conclusion that there is
no association of BDNF rs6265 and rs12273363 with MA and/or MO. For rs2049046 the increase in
frequency of the TT genotype or T-allele for the combined cohort 1 and 2 migraine or MA cases
compared to controls shows significance.
11
BDNF has an important role in a number of aspects of neuronal development and synaptic function
and variants in BDNF have been associated with a wide array of diseases and conditions reflecting its
involvement in neuronal function. Because of the correlation of BDNF levels with a number of aspects
related to migraine studies have previously investigated some BDNF SNPs in relation to migraine risk.
We find no association of migraine with the SNPs rs151948, rs7127507 and rs12273363, which had
not been tested with respect to migraine previously, but had shown positive associations with other
neurological conditions, such as bipolar and mood disorders [35, 36]. We also investigated two SNPs
(rs6265 and rs2049046) that have been tested previously in other migraine populations, however, in
this study, we have used a larger population which increases power, reducing the likelihood of false
negatives, and we also have a greater proportion of migraineurs with a MA diagnosis compared to
MO, increasing the likelihood of detecting an effect in this migraine sub-type, which we hypothesized
BDNF may be more relevant to. Similar to the studies by Marziniak et al. (2008) and Lemos et al. (2010)
we find no evidence of the functional BDNF SNP rs6265 as a risk factor for migraine, nor the migraine
subtypes MO and MA. Although rs6265 is the most extensively studied SNP in the BDNF locus because
it results in a Val to Met amino acid change that is known to affect function of the protein, we did not
find that an increased prevalence of the Met/Met genotype or Met allele in our migraine population
and more specifically in MA cases, which is consistent with other studies with a larger MO component.
The rs6265 SNP is within the pro-protein region of BDNF and affects the level of mature protein [29],
so the fact that there is no association of this SNP with migraine suggests that reduced BDNF protein
levels in the brain do not increase the likelihood of migraine. The rs7127507 and rs12273363 SNPs
have also been shown to influence pro-BDNF protein levels; the minor allele in each case has been
shown to be associated with a decrease of pro-BDNF density in the hippocampus in post-mortem
brains [37]. rs12273363 resides in promoter IV, which directs transcription of the most 5’ transcripts
in the hippocampus, cortex and amygdala, modulating its activity in an allele-specific manner [35].
We initially detected a significant difference in genotype and allele frequencies for the rs2049046 SNP
12
decrease in the percentage of individuals with the rs2049046 AA-genotype and an increase in those
with the TT-genotype in migraineurs, this was not significant for either total migraine or with respect
to the MO or MA sub-groups. Lemos et al. (2010) had found an increased risk of migraine associated
with the heterozygous AT genotype at rs2049046, but only in association with the heterozygous
genotype the calcitonin gene-related peptide SNP rs1553005 [31]. The rs2049046 SNP is located at
the 5’ end of the BDNF locus, in promoter III upstream of some of the alternative transcription start
sites of the gene and a region that also contains SNPs that have been identified from genome wide
association studies for obesity [26]. It has been shown that with respect to obesity, it is the absence
of a specific transcript of BDNF with a long 3’UTR that is causal rather than the level of BDNF mRNA in
a mouse obesity syndrome model [38]. Conceivably, similar to rs7127507 and rs12273363, SNPs in the
5’ end of the locus may also influence tissue-specific transcription or levels of particular BDNF
transcripts for obesity, as well as any other conditions that show associations.
In the population studied here we found high LD (calculated by D’) throughout the BDNF locus with
most of the gene encompassed in a single block (Fig. 1b). This agrees with data from Hapmap (Fig. 1a)
and other BDNF SNP association studies of populations with conditions such as bipolar disorder, opoid
addiction and Alzheimer’s disease [21, 23, 36]. These data sets all found high LD across the locus, but
fine mapping revealed a few interspersed local regions of lower LD with independent SNPs. This may
reflect the important function of the gene in many aspects of brain function as well as the complicated
regulatory landscape of the locus. Minor allele frequencies were similar to those reported for the CEU
population in HapMap. However, the allelic frequencies for all the BDNF SNPs investigated in this study
appear to vary greatly between the different Hapmap populations, so conclusions from one
population may not apply in another population and care would need to be taken that ethnicity is
taken into account.
It should be noted that SNPs at the BDNF locus have not reached genome-wide significance in the
genome-wide association studies that have been performed for common migraine (which included
13
to date only a small number of genes have been found to be robustly implicated in migraine [41]. In
conclusion, we found that rs2049046, which resides at the 5’ end of one the BDNF transcripts, to be
significantly associated with migraine, and in particular MA, in an Australian migraine case-control
population. While a second cohort showed a similar trend, the results were not significant. Thus while
there is no conclusive evidence to implicate the BDNF rs2049046 genotype in migraine susceptibility,
larger studies may help to either rule it in or out if the effect size is low.
14
Figure 1. Gene transcription, SNP location and haploblock structure of the BDNF locus. A. Schematic
of the the SNPs genotyped in this study in relationship to the BDNF gene and some of its transcripts,
and linkage disequilibrium (LD) values between SNPs from Hapmap data at the locus. Top panel: SNPs
genotyped in this study are boxed in yellow with arrows pointing to position along the BDNF locus.
BDNF isoforms are transcribed from the reverse strand (direction indicated by dashed arrow) and a
subset are depicted to indicate the main regions of initiation (see [7] for a full description, including
antisense transcripts). Black boxes represent the protein coding sequence, while dark grey boxes
represent untranslated exons. Bottom panel: Hapmap data was used to generate the LD values
(calculated by D’) for multiple SNPs across the BDNF locus from rs1519480 (chr11:27675462) to
rs2049048 (chr11:27750836) in Haploview, including rs1519480, rs6265, rs712507 and rs12273363.
rs2049046 was not available in Hapmap.. Darker squares indicated higher LD; dark red squares
indicate SNPs are in complete LD (100% or D’ =1), and numbers in lighter squares are the percentage
when LD is incomplete. B. LD values generated from the BDNF SNPs genotyped in Cohort 1 from this
15
Acknowledgements: We would like to thank all the participants of the study. This research was
supported by funding from an Australian DEST International Science Linkages grant and by
infrastructure purchased with Australian Government EIF Super Science Funds as part of the
Therapeutic Innovation Australia - Queensland Node project. Astrid J. Rodriguez-Acevedo was the
recipient of a Griffith University International Postgraduate Research Award.
REFERENCES
1. Lipton RB, Bigal ME, Diamond M, Freitag F, Reed ML, Stewart WF. Migraine prevalence,
disease burden, and the need for preventive therapy. Neurology. 2007;68:343-349.
2. Headache Classification Committee of the International Headache Society. The international
Classification of the Headache Disorders, 3rd ed. (beta version). Cephalalgia.
16
3. Olesen J, Goadsby, J.G., Ramadan, N.H., Tfelt-Hansen, P. and Welch, K.M.H. (Eds.). The
Headaches third ed. 3rd edn: Lippincott Williams & Wilkins, Philadelphia; 2006.
4. Lipsky RH, Marini AM. Brain-derived neurotrophic factor in neuronal survival and
behavior-related plasticity. Ann N Y Acad Sci. 2007;1122:130-143.
5. Thompson SW, Bennett DL, Kerr BJ, Bradbury EJ, McMahon SB. Brain-derived neurotrophic
factor is an endogenous modulator of nociceptive responses in the spinal cord. Proc Natl Acad
Sci U S A. 1999;96:7714-7718.
6. Mannion RJ, Costigan M, Decosterd I, Amaya F, Ma QP, Holstege JC, Ji RR, Acheson A, Lindsay
RM, Wilkinson GA, Woolf CJ. Neurotrophins: peripherally and centrally acting modulators of
tactile stimulus-induced inflammatory pain hypersensitivity. Proc Natl Acad Sci U S A.
1999;96:9385-9390.
7. Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus:
bidirectional transcription, complex splicing, and multiple promoters. Genomics.
2007;90:397-406.
8. Seidah NG, Benjannet S, Pareek S, Chretien M, Murphy RA Cellular processing of the
neurotrophin precursors of NT3 and BDNF by the mammalian proprotein convertases. FEBS
Lett. 1996;379:247-250.
9. Lu B, Pang PT, Woo NH. The yin and yang of neurotrophin action. Nat Rev Neurosci.
2005;6:603-614.
10. Mossner R, Mikova O, Koutsilieri E, Saoud M, Ehlis AC, Muller N, Fallgatter AJ, Riederer P.
Consensus paper of the WFSBP Task Force on Biological Markers: biological markers in
depression. World J Biol Psychiatry. 2007;8:141-174.
11. Terracciano A, Piras MG, Lobina M, Mulas A, Meirelles O, Sutin AR, Chan W, Sanna S, Uda M,
Crisponi L, Schlessinger D. Genetics of serum BDNF: meta-analysis of the Val66Met and
17
12. Tanure MT, Gomez RS, Hurtado RC, Teixeira AL, Domingues RB. Increased serum levels of
brain-derived neurotropic factor during migraine attacks: a pilot study. J Headache Pain.
2010;11:427-430.
13. Fischer M, Wille G, Klien S, Shanib H, Holle D, Gaul C, Broessner G. Brain-derived neurotrophic
factor in primary headaches. J Headache Pain. 2012;13:469-475.
14. Blandini F, Rinaldi L, Tassorelli C, Sances G, Motta M, Samuele A, Fancellu R, Nappi G, Leon A.
Peripheral levels of BDNF and NGF in primary headaches. Cephalalgia. 2006;26:136-142.
15. Pietrobon D. Migraine: new molecular mechanisms. Neuroscientist. 2005.11:373-386.
16. Buldyrev I, Tanner NM, Hsieh HY, Dodd EG, Nguyen LT, Balkowiec A Calcitonin gene-related
peptide enhances release of native brain-derived neurotrophic factor from trigeminal
ganglion neurons. J Neurochem. 2006;99:1338-1350.
17. Urbach A, Bruehl C, Witte OW Microarray-based long-term detection of genes differentially
expressed after cortical spreading depression. Eur J Neurosci. 2006;24:841-856.
18. Bolay H, Reuter U, Dunn AK, Huang Z, Boas DA, Moskowitz MA. Intrinsic brain activity triggers
trigeminal meningeal afferents in a migraine model. Nat Med. 2002;8:136-142.
19. Zhang X, Levy D, Noseda R, Kainz V, Jakubowski M, Burstein R. Activation of meningeal
nociceptors by cortical spreading depression: implications for migraine with aura. J Neurosc.i
2010;30:8807-8814.
20. Mandelman SD, Grigorenko EL. BDNF Val66Met and cognition: all, none, or some? A
meta-analysis of the genetic association. Genes Brain Behav. 2012;11:127-136.
21. Honea RA, Cruchaga C, Perea RD, Saykin AJ, Burns JM, Weinberger DR, Goate AM, Alzheimer's
Disease Neuroimaging I. Characterizing the role of brain derived neurotrophic factor genetic
variation in Alzheimer's disease neurodegeneration. PLoS One. 2013;8:e76001.
22. Hall D, Dhilla A, Charalambous A, Gogos JA, Karayiorgou M. Sequence variants of the
brain-derived neurotrophic factor (BDNF) gene are strongly associated with obsessive-compulsive
18
23. Liu L, Foroud T, Xuei X, Berrettini W, Byerley W, Coryell W, El-Mallakh R, Gershon ES, Kelsoe
JR, Lawson WB, et al. Evidence of association between brain-derived neurotrophic factor gene
and bipolar disorder. Psychiatr Genet. 2008;18:267-274.
24. Zhang H, Ozbay F, Lappalainen J, Kranzler HR, van Dyck CH, Charney DS, Price LH, Southwick
S, Yang BZ, Rasmussen A, Gelernter J. Brain derived neurotrophic factor (BDNF) gene variants
and Alzheimer's disease, affective disorders, posttraumatic stress disorder, schizophrenia, and
substance dependence. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:387-393.
25. Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A,
Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, et al. Genome-wide association
yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet.
2009;41:18-24.
26. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren
CM, Luan J, Magi R, et al. Association analyses of 249,796 individuals reveal 18 new loci
associated with body mass index. Nat Genet 2010;42:937-948.
27. Jiao H, Arner P, Hoffstedt J, Brodin D, Dubern B, Czernichow S, van't Hooft F, Axelsson T,
Pedersen O, Hansen T, et al. Genome wide association study identifies KCNMA1 contributing
to human obesity. BMC Med Genomics. 2011;4:51.
28. Okada Y, Kubo M, Ohmiya H, Takahashi A, Kumasaka N, Hosono N, Maeda S, Wen W, Dorajoo
R, Go MJ, et al Common variants at CDKAL1 and KLF9 are associated with body mass index in
east Asian populations. Nat Genet. 2012, 44:302-306.
29. Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B,
Goldman D, Dean M, et al. The BDNF val66met polymorphism affects activity-dependent
secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257-269.
30. Marziniak M, Herzog AL, Mossner R, Sommer C. Investigation of the functional brain-derived
neurotrophic factor gene variant Val66MET in migraine. J Neural Transm.
19
31. Lemos C, Mendonca D, Pereira-Monteiro J, Barros J, Sequeiros J, Alonso I, Sousa A: BDNF and
CGRP interaction: implications in migraine susceptibility. Cephalalgia 2010, 30:1375-1382.
32. Colson NJ, Lea RA, Quinlan S, MacMillan J, Griffiths LR. The estrogen receptor 1 G594A
polymorphism is associated with migraine susceptibility in two independent case/control
groups. Neurogenetics. 2004;5:129-133.
33. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from
human nucleated cells. Nucleic Acids Res. 1988;16:1215.
34. Yogeetha BS, Haupt LM, McKenzie K, Sutherland HG, Okolicsyani RK, Lea RA, Maher BH, Chan
RC, Shum DH, Griffiths LR. BDNF and TNF-alpha polymorphisms in memory. Mol Biol Rep.
2013;40:5483-5490.
35. Hing B, Davidson S, Lear M, Breen G, Quinn J, McGuffin P, MacKenzie A. A polymorphism
associated with depressive disorders differentially regulates brain derived neurotrophic factor
promoter IV activity. Biol Psychiatry. 2012;71:618-626.
36. de Cid R, Fonseca F, Gratacos M, Gutierrez F, Martin-Santos R, Estivill X, Torrens M. BDNF
variability in opioid addicts and response to methadone treatment: preliminary findings.
Genes Brain Behav. 2008;7:515-522.
37. Dunham JS, Deakin JF, Miyajima F, Payton A, Toro CT. Expression of hippocampal
brain-derived neurotrophic factor and its receptors in Stanley consortium brains. J Psychiatr Res.
2009;43:1175-1184.
38. Liao GY, An JJ, Gharami K, Waterhouse EG, Vanevski F, Jones KR, Xu B. Dendritically targeted
Bdnf mRNA is essential for energy balance and response to leptin. Nat Med. 201218:564-571.
39. Chasman DI, Schurks M, Anttila V, de Vries B, Schminke U, Launer LJ, Terwindt GM, van den
Maagdenberg AM, Fendrich K, Volzke H, et al. Genome-wide association study reveals three
susceptibility loci for common migraine in the general population. Nat Genet.
20
40. Freilinger T, Anttila V, de Vries B, Malik R, Kallela M, Terwindt GM, Pozo-Rosich P, Winsvold B,
Nyholt DR, van Oosterhout WP, et al. Genome-wide association analysis identifies
susceptibility loci for migraine without aura. Nat Genet. 2012;44:777-782.
41. Jones B. Migrane: new susceptibility loci associated with migraine. Nat Rev Neurol.
21
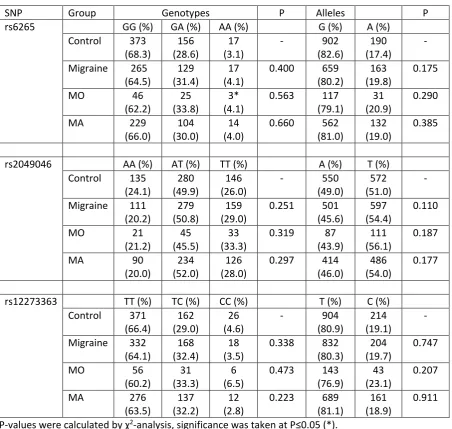
Table 1 . Genotype and allele frequencies for BDNF SNPs in migraine case-control cohort 1
SNP Group Genotypes P Alleles P
rs1519480 TT (%) TC (%) CC (%) T (%) C (%) Control 121
(51.3)
94 (39.8)
21 (8.9)
- 336 (71.2)
136 (28.8)
-
Migraine 115 (48.7)
96 (40.7)
25 (10.6)
0.771 326 (69.1)
146 (30.9)
0.755
MO 50
(55.6)
34 (37.8)
6 (6.7)
0.708 134 (74.4)
46 (25.6)
0.407
MA 65
(44.5)
62 (42.5)
19 (13.0)
0.293 192 (65.8)
100 (34.2)
0.114
rs6265 GG (%) GA (%) AA (%) G (%) A (%)
Control 171 (69.5)
68 (27.6)
7 (2.8)
- 410 (83.3)
82 (16.7)
-
Migraine 131 (65.2)
64 (31.8)
6 (3.0)
0.614 326 (81.1)
76 (18.9)
0.383
MO 49
(70.0)
19 (27.1)
2 (2.9)
NC 117 (83.6)
23 (16.4)
0.947
MA 82
(62.6)
45 (34.4)
4 (3.1)
NC 209 (79.8)
53 (20.2)
0.224
rs7127507 TT (%) TC (%) CC (%) T (%) C (%) Control 114
(46.2)
113 (45.7)
20 (8.1)
- 341 (69.0)
153 (31.0)
-
Migraine 114 (46.3)
108 (43.9)
24 (9.8)
0.789 336 (68.3)
156 (31.7)
0.803
MO 49
(51.6)
39 (41.1)
7 (7.4)
0.667 137 (72.1)
53 (27.9)
0.432
MA 65
(43.0)
69 (45.7)
17 (11.3)
0.546 199 (65.9)
103 (34.2)
0.358
rs2049046 AA (%) AT (%) TT (%) A (%) T (%) Control 60
(24.6)
128 (52.5)
56 (23.0)
- 248 (50.8)
240 (49.2)
-
Migraine 46 (19.6)
115 (48.9)
74 (31.5)
0.088 207 (44.0)
263 (56.0)
0.035*
MO 23
(27.1)
36 (42.4)
26 (32.0)
0.232 82 (48.2)
88 (51.8)
0.562
MA 23
(15.3)
79 (52.7)
48 (29.0)
0.036* 125 (41.7)
175 (58.3)
0.0125*
rs12273363 TT (%) TC (%) CC (%) T (%) C (%) Control 167
(66.0)
74 (29.2)
12 (4.7)
- 408
(80.6)
98 (19.4)
-
Migraine 160 (64.5)
79 (31.9)
9 (3.6)
0.708 339 (68.3)
97 (19.6)
0.940
MO 70
(72.9)
25 (26.0)
1 (1.0)
NC 165 (85.9)
27 (14.1)
22
MA 90
(59.2)
54 (35.5)
8 (5.3)
0.382 234 (77.0)
70 (23.0)
0.214
P-values were calculated by χ2-analysis, significance was taken at P≤0.05 (*).
NC p-value not calculated as minimum number for each category needs to be >5 for χ2-squared
analysis.
23
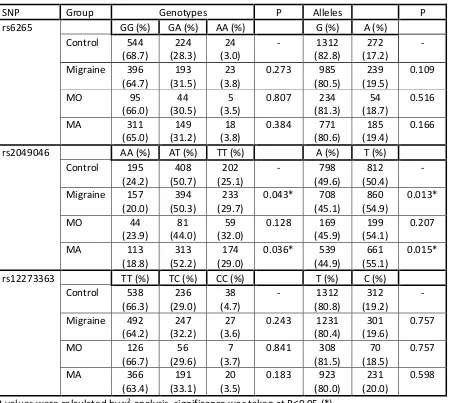
Table 2. Genotype and allele frequencies for BDNF SNPs in migraine case-control cohort 2
SNP Group Genotypes P Alleles P
rs6265 GG (%) GA (%) AA (%) G (%) A (%)
Control 373 (68.3)
156 (28.6)
17 (3.1)
- 902 (82.6)
190 (17.4)
-
Migraine 265 (64.5)
129 (31.4)
17 (4.1)
0.400 659 (80.2)
163 (19.8)
0.175
MO 46
(62.2)
25 (33.8)
3* (4.1)
0.563 117 (79.1)
31 (20.9)
0.290
MA 229
(66.0)
104 (30.0)
14 (4.0)
0.660 562 (81.0)
132 (19.0)
0.385
rs2049046 AA (%) AT (%) TT (%) A (%) T (%) Control 135
(24.1)
280 (49.9)
146 (26.0)
- 550 (49.0)
572 (51.0)
-
Migraine 111 (20.2)
279 (50.8)
159 (29.0)
0.251 501 (45.6)
597 (54.4)
0.110
MO 21
(21.2)
45 (45.5)
33 (33.3)
0.319 87 (43.9)
111 (56.1)
0.187
MA 90
(20.0)
234 (52.0)
126 (28.0)
0.297 414 (46.0)
486 (54.0)
0.177
rs12273363 TT (%) TC (%) CC (%) T (%) C (%) Control 371
(66.4)
162 (29.0)
26 (4.6)
- 904 (80.9)
214 (19.1)
-
Migraine 332 (64.1)
168 (32.4)
18 (3.5)
0.338 832 (80.3)
204 (19.7)
0.747
MO 56
(60.2)
31 (33.3)
6 (6.5)
0.473 143 (76.9)
43 (23.1)
0.207
MA 276
(63.5)
137 (32.2)
12 (2.8)
0.223 689 (81.1)
161 (18.9)
0.911
P-values were calculated by χ2-analysis, significance was taken at P≤0.05 (*).
NC p-value not calculated as minimum number for each category needs to be >5 for χ2-squared
analysis.
24
Table 3. Genotype and allele frequencies for BDNF SNPs in migraine case-control population of cohorts 1 and 2 combined.
SNP Group Genotypes P Alleles P
rs6265 GG (%) GA (%) AA (%) G (%) A (%)
Control 544 (68.7)
224 (28.3)
24 (3.0)
- 1312 (82.8)
272 (17.2)
-
Migraine 396 (64.7)
193 (31.5)
23 (3.8)
0.273 985 (80.5)
239 (19.5)
0.109
MO 95
(66.0)
44 (30.5)
5 (3.5)
0.807 234 (81.3)
54 (18.7)
0.516
MA 311
(65.0)
149 (31.2)
18 (3.8)
0.384 771 (80.6)
185 (19.4)
0.166
rs2049046 AA (%) AT (%) TT (%) A (%) T (%) Control 195
(24.2)
408 (50.7)
202 (25.1)
- 798 (49.6)
812 (50.4)
-
Migraine 157 (20.0)
394 (50.3)
233 (29.7)
0.043* 708 (45.1)
860 (54.9)
0.013*
MO 44
(23.9)
81 (44.0)
59 (32.0)
0.128 169 (45.9)
199 (54.1)
0.207
MA 113
(18.8)
313 (52.2)
174 (29.0)
0.036* 539 (44.9)
661 (55.1)
0.015*
rs12273363 TT (%) TC (%) CC (%) T (%) C (%) Control 538
(66.3)
236 (29.0)
38 (4.7)
- 1312 (80.8)
312 (19.2)
-
Migraine 492 (64.2)
247 (32.2)
27 (3.6)
0.243 1231 (80.4)
301 (19.6)
0.757
MO 126
(66.7)
56 (29.6)
7 (3.7)
0.841 308 (81.5)
70 (18.5)
0.757
MA 366
(63.4)
191 (33.1)
20 (3.5)
0.183 923 (80.0)
231 (20.0)
0.598
P-values were calculated by χ2-analysis, significance was taken at P≤0.05 (*).