The
archiving
and
dissemination
of
biological
structure
data
Helen
M
Berman
1,
Stephen
K
Burley
1,2,
Gerard
J
Kleywegt
3,
John
L
Markley
4,
Haruki
Nakamura
5and
Sameer
Velankar
3TheglobalProteinDataBank(PDB)wasthefirstopen-access
digitalarchiveinbiology.ThehistoryandevolutionofthePDB
aredescribed,togetherwiththewaysinwhichmolecular
structuralbiologydataandinformationarecollected,curated,
validated,archived,anddisseminatedbythemembersofthe
WorldwideProteinDataBankorganization(wwPDB;http://
wwpdb.org).Particularemphasisisplacedontheroleof
communityinestablishingthestandardsandpoliciesbywhich
thePDBarchiveismanagedday-to-day.
Addresses 1
ResearchCollaboratoryforStructuralBioinformaticsProteinData Bank,DepartmentofChemistryandChemicalBiology,Centerfor IntegrativeProteomicsResearch,InstituteforQuantitativeBiomedicine, Rutgers, The State University of New Jersey, 174 Frelinghuysen Road, Piscataway, NJ 08854, USA
2
ResearchCollaboratoryforStructuralBioinformaticsProteinData Bank,SkaggsSchoolofPharmacyandPharmaceuticalSciencesand SanDiegoSupercomputerCenter,UniversityofCaliforniaSanDiego, 9500 Gilman Drive, La Jolla, CA 92093, USA
3Protein Data Bank in Europe, European Molecular Biology Laboratory—EuropeanBioinformaticsInstitute,WellcomeGenome Campus,CambridgeCB101SD,UK
4BiologicalMagneticResonanceBank,DepartmentofBiochemistry, University of Wisconsin-Madison, Madison, WI 53706, USA 5Protein Data Bank Japan, Institute for Protein Research, Osaka University,3-2Yamadaoka,Suita,Osaka,565-0871,Japan Correspondingauthor:Berman,HelenM(berman@rcsb.rutgers.edu)
CurrentOpinioninStructuralBiology2016, 40:17–22 This review comes from a themed issue on Biophysicaland molecularbiologicalmethods
Edited by PetraFrommeand AndrejSali
http://dx.doi.org/10.1016/j.sbi.2016.06.018
0959-440/#2016TheAuthors.PublishedbyElsevierLtd.Thisisan openaccessarticleundertheCCBY-NC-NDlicense( http://creative-commons.org/licenses/by-nc-nd/4.0/)
Historical
background
Structural biologyis arelativelyyoung sciencethatcan
trace its roots to the first X-ray diffraction studies of
pepsin in 1935 by Dorothy Crowfoot (Hodgkin), who
at the time was a student of J.D. Bernal [1]. Twenty
years later, Kendrew determined thestructure of
myo-globin [2,3]; shortly thereafter, Perutz determined the
structureofhemoglobin[4,5].BothwonNobelprizesfor
theirachievements.Notlongafterthesestructureswere
published,thecrystallographiccommunitybegan
discus-sionsastohowtobestarchivethesedataandmakethem
available.Duringthisperiod,therewerenumerous
grass-rootsmeetings,one ofwhich resultedin apetition,and
manyexchangesofhandwrittendocuments.In1971,the
ColdSpring HarborLaboratoryhosted asymposium on
proteincrystallography,duringwhichleadersinthefield
presented theirseminal work[6].Walter Hamilton,an
attendee,offeredtoprovidethefirsthomeforwhatisnow
knownastheProteinDataBank(PDB)[7].ThePDBwas
launched at Brookhaven National Laboratory, on the
basisof theProteinStructure LibrarycreatedbyEdgar
Meyer[8].TheinitialPDBarchivecontainedfewerthan
ten structures, all of which were determined by X-ray
crystallography.Inthe1980s,structuresdeterminedusing
NMR methodsbegan to bedeposited, andin 1990 the
first structure determined by electron microscopy was
deposited. In 1982 the PDB reached 100 entries, in
1993 1000 entries, in 1999 10000, and in 2014
100000entries.Atthetimeofwriting,thePDBarchive
containsover117000structuresofproteins,nucleicacids,
and their complexes with one another and with small
molecule ligands.
The
PDB
as
a
community
data
resource
Fromitsinception,thePDBhasbeenacommunityeffort
that hasevolved with changesin scientificculture.For
example,whenthePDBwasfirst created,data
submis-sionwas voluntary.However,inthe1980s, membersof
the community became outspoken about the need to
enforce mandatory datadeposition. Various committees
weresetup todefinewhatdatashould berequiredand
when to disseminate the data. These guidelines were
publishedin1989,andovertime,adoptedbyvirtuallyall
of the scientificjournals thatnow requirePDB
deposi-tion(s) as a prerequisite for publication of structural
studies [9].In 2008, further shifts in community
senti-ment ledto mandatory deposition of experimentaldata
togetherwithatomiccoordinates.Inthecurrentdecade,
theimportanceof reproducibility hasbeen highlighted.
The PDB convened method-specific Validation Task
Forces and Workshops [10,11,12,13] to define
whatdatashould becollected and how best to validate
thestructuralmodels,theexperimentaldata,andthefitof
themodelstothedata.NoweverystructureinthePDB
authorsarestronglyencouragedtoincludethesereports
withtheirmanuscriptsubmissionstojournals.
Theimportanceofglobal participationindataarchiving
wasunderstoodearlyinthecreationofthePDB.Indeed,
the announcement of the PDB in 1971 described the
collaborationwiththeCambridgeCrystallographic
Data-baseCentre[7].In2003,aMemorandumof
Understand-ing (MOU) among partners in the US (RCSB Protein
Data Bank; http://www.rcsb.org), Japan (Protein Data
BankJapan orPDBj; http://www.pdbj.org),and Europe
(ProteinDataBankinEuropeorPDBe;http://pdbe.org)
establishedtheWorldwideProteinDataBank(wwPDB)
partnership, which is responsible for formalizing the
proceduresinvolvedincollecting,standardizing,
annotat-ing and disseminating the data [14]. Subsequently, a
globalNMR specialistdatarepositoryBioMagResBank,
composedof deposition sites in theUS(BMRB;http://
www.bmrb.wisc.edu) and Japan (PDBj-BMRB; http:// bmrbdep.pdbj.org),joinedthewwPDB.
TheX-raycrystallographycommunityhasledthe
biolog-ical sciences in the area of data sharing. While the
sociological/anthropologicalunderpinningsofthis
leader-shiprolehavenotbeenfullyexplored,muchofwhathas
transpiredin thecreationandevolutionof thePDBcan
be traced to J.D. Bernal, who, in addition to being a
brilliant scientificinnovator,was a prominent social
ac-tivist,whosebeliefswereconsistentwiththeconductof
thePDB[15].
Content
of
the
PDB
archive
ThePDBarchivecontainsinformationaboutstructural
models that have been derived from experimental
methods,includingX-ray/neutron/electron
crystallogra-phy, NMR spectroscopy, and 3D electron microscopy
(3DEM).Inadditiontothe3Dcoordinates,thedetails
ofthe chemistry ofthe polymers and small molecules
arearchived,asaremetadatadescribingthe
experimen-tal conditions, data-processing statistics and structural
featuressuchasthesecondaryandquaternarystructure.
Thestructure-factoramplitudes(orintensities)usedto
determine X-ray structures, and chemical shifts and
restraintsusedindeterminingNMRstructuresare also
archived. The electron density maps used to derive
3DEM models are archived in EMDB [16], and the
experimentaldata underpinningthemcan bearchived
in EMPIAR [17]. In collaboration with community
experts, pertinent data items are defined for each
experimental field, with requirements evolving over
time. The PDB data dictionary, originally developed
to describe macromolecular crystallography, contains
more than 4400 data items. The dictionary combines
dataitemscommontoallmethodsaswellasthosethat
are method specific. For example, the current
dictio-narycontains 250NMR-specificdataand1200
3DEM-specific data definitions.
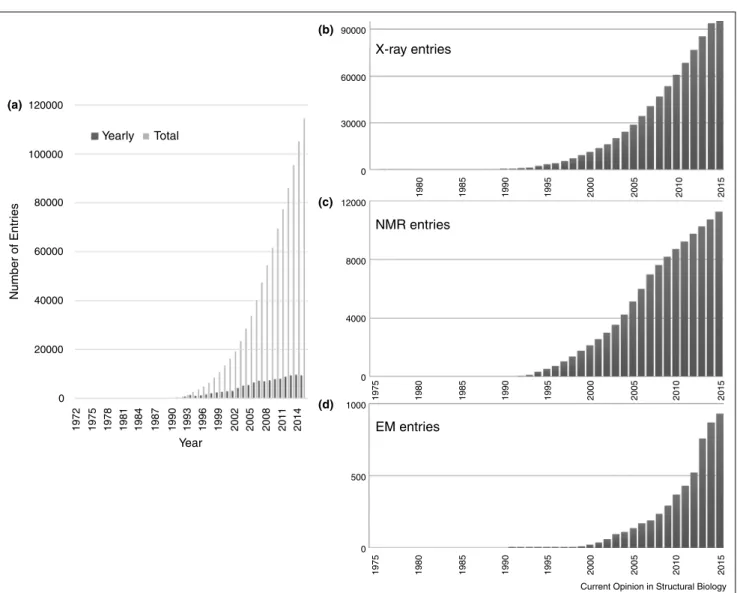
Over time, the holdings of the PDB have increased
dramaticallyashasthecomplexityofthestructuresbeing
archived(Figure1).
Aworkshopheldin2005ledtothepolicythatpurelyinsilico
modelsshouldnotbepartofthePDB[18],and,instead,a
modelingportalshouldbecreatedforthesemodels.The
ProteinModelingPortalwasestablishedin2007[19].
Representation
of
PDB
data
ThefirstdataformatusedbythePDBwasestablishedin
theearly 1970s and was onthe basis of the 80-column
Hollerith format used for punched cards. The atom
recordsincludedatomname,residuenameandsequence
number. A ‘header record’ contained some metadata.
Thisformatwasreadily acceptedbecauseitwas simple
andbothhuman-andmachine-readable.However,ithad
manyseriousdrawbacksinthatthesizeofthestructural
modelswaslimitedto99999atomsandthatrelationships
among the data items were implicit. These inherent
weaknesses meant that significant domain knowledge
wasnecessaryinordertowritesoftwareusingthisformat.
Inthe1990s,theIUCrcharteredacommitteetocreatea
more formaldata model.This committee proposedthe
Macromolecular Crystallographic Information File
(mmCIF)[20].mmCIFisaself-definingformatinwhich
every data item has attributes describing its features
includingrelationshipstootherdataitems.Most
impor-tantly,mmCIFhasnolimitationswithrespecttothesize
ofthearchivedstructuralmodel.Thedictionaryandthe
datafiles are completely machine-readable,and no
do-mainknowledge is required to read thefiles. The first
dictionarycontainedover3000dataitemsrelevantto
X-ray crystallography. Over time, terms specific to NMR
and3DEMwereadded,andthedictionarywasrenamed
PDBx/mmCIF.In2007,itwasdecidedthatPDBxwould
betheMasterFormatfordatacollectedbythePDB.In
2011,majorX-raystructuredeterminationsoftware
devel-opersagreedto adoptthis datamodelso thatalloutput
from theirprograms wouldbe in PDBx. In 2015,large
structures archivedin the PDBthat had formerly been
split into multiple entries were combined into single
entries and mmCIF formatted files. Other structural
biology communities are in the process of building on
the PDBx/mmCIF framework to establish their own
controlledvocabularyandspecialist dataitems[19,21].
PDBML,anXMLformatonthebasisofPDBx/mmCIF
[22], and its RDF (Resource Description Framework)
conversionweredevelopedtofacilitatetheintegrationof
structure data with other life sciences data resources
couldbefacilitated [23].
The
data
pipeline
Everydataresourcehasasetofproceduresfordeposition,
ThepipelinecurrentlyusedbythewwPDBtopopulate
thePDBarchiveisillustratedschematicallyinFigure2.
In the very early days of the PDB, structures were
deposited toBNL onmagnetictapescontainingatomic
coordinateswithpaperformslistingotherdataitems,all
sentfirstbymailandthenviaAweb-basedsystem,called
AutoDep,wascreatedinthe1990s[24].Thissystemwas
latermodifiedandusedbyPDBe[25]untilveryrecently.
TheRCSBPDBandPDBjcollecteddatausingasystem
onthebasisofmmCIFcalledADIT[26],andtheBMRB
in the US and its affiliate in Japan adopted a similar
systemcalledADIT-NMR[27].Althoughthesesystems
were distinct, since 2003, the wwPDB partners have
determined jointly what data should be collected and
whichproceduresandalgorithmsshouldbeusedfordata
processing.In2007,itwasagreedwithinthewwPDBto
createasingledeposition,Structuresaremadeavailable
to the public either immediately after they have been
fullycuratedor-inmostcases-whentheyarepublishedin
ajournal.Usually,eithertheauthororthejournalinforms
wwPDBthatthepaperdescribingthestructureisaboutto
be published. PDB data are released in a two-stage
process. Every Saturday at 03:00 UTC the polymer
sequences, ligand SMILES strings, and crystallization
pH for new structures designated for release are made
public(http://wwpdb.org/download/downloads)asa
cour-tesytotheproteinstructuremodelingandcomputational
chemistrycommunitiestoenableweeklyblinded
predic-tion challenge efforts (e.g., CAMEO [19] and D3R
CELPP [28]). Every Wednesday at 00:00 UTC, all
new structuresdesignatedfor releasearemadepublicly
available through the wwPDB FTP sites. On average
about200structuresarereleasedeveryweek.Asevidence
Figure1 120000 (a) (b) (c) (d) 100000 80000 60000 40000 20000 0 1972 1975 1978 1981 1984 1987 1990 1993 1996 1999 2002 2005 2008 2011 2014 Year Number of Entr ies Yearly Total 90000 60000 X-ray entries NMR entries EM entries 30000 0 12000 8000 4000 1000 500 0 0 1980 1985 1990 1995 2000 2005 2010 2015 1980 1975 1985 1990 1995 2000 2005 2010 2015 1980 1975 1985 1990 1995 2000 2005 2010 2015
Current Opinion in Structural Biology
GrowthofthePDBarchive.(a)Numberofentriesdepositedannually(darkgray)andavailableattheendofeachyear(lightgray);(b)numberof X-ray crystal structures; (c)NMR structures, and D) 3DEM structures available each year.
for the importance of this archive, in 2015, more than
500millionsetsofatomiccoordinatesweredownloaded
fromthewwPDB FTPsites.
Value-added
resources
ThewwPDB FTPsitesprovidethecoredataformany
databases,services,andwebsites,includingthoserunby
theindividualwwPDBpartners.IntheoriginalwwPDB
MOU,itwasagreedthattobest servescience,wwPDB
partner websites would compete with one another and
wouldoffermanydifferentkindsofservicesandfeatures.
The RCSB PDB has extensive search and reporting
capabilities as well as an education portal called
PDB-101 [26,29]. PDBe has multiple search and browse
facilities as well as analysis and bioinformatics tools
[30,31].PDBjprovidesavarietyofservicesandviewers
and supports browsing in multiple Asian languages
[23,32].BMRBhasmanycapabilitiesdesignedtoserve
theNMRcommunity[33].
CATH[34]andSCOP[35,36]usethedatainthePDBto
classifythestructuraldomainsofproteinswithanattempt
to relate them to function. More recently, these two
databaseshave agreed to worktogether and with other
resourcesintheUK toprovidepredictedstructural
fea-turesunder aunifiedsystemcalled Genome3D[37].
Additional specialty databases provide information on
particular classes of macromolecules such as nucleic
acids[38].
TheProteinStructureInitiative(PSI)StructuralBiology
Knowledgebase (SBKB)[39]was an ambitiouseffort to
unifyinformationaboutproteinsequence, structureand
function. Unfortunately, the decision to discontinue
fundingthe PSI meansthat this resource will cease to
exist.
Challenges
going
forward
A review of the holdings of the PDB shows a steady
growth(10,000new structuresannually).More
signifi-cantly,thecomplexityofthestructuralmodelscontinues
to increase with more and more large heterogeneous
assemblies entering the archive. Fortunately, there are
nolongertechnicalrestrictionsto receiving,annotating,
validating,anddisseminatingtheseverylargestructures.
Historically,moststructuresweredeterminedexclusively
with the aid of a single experimental method: X-ray
crystallography, NMRor 3DEM. Inrecentyears, these
traditional techniques are being combined with other
methods to yield improved models. For example, it is
nowcommonpracticetoadddatafromsmall-angle
scat-tering measurements to NMR-derived restraints to
de-terminesolutionstructures[40,41].Similarly,NMRor
X-raydatacanbecombinedwithcryoEMdatainintegrative
modeling approaches [42]. Such integrative methods
makeitpossibletocombinedatafromdifferent
biophys-ical techniques with computational methods to create
models of very large macromolecular machines [43].
However, hybrid approaches also present a variety of
challenges including how to validate these structures
andthen howto archivethem. Asin thepast, withthe
help and advice of an expert Task Force [44], this
integrativechallengewillbemetbythewwPDBpartners.
Figure2
Deposition Pipeline
Format conversion
and data harvesting Validation Ligand Lite Communication
Workflow-Automation System Release Processing Deposition Interface: Annotation Pipeline X-ray-specific a. EM-specific b. NMR-specific c. Communication Report generation/ Status Tracking Validation Added Annotations Sequence Processing Ligand Processing
Current Opinion in Structural Biology
Acknowledgements
RCSBPDBissupportedbyNSF[DBI-1338415],NIH,DOE;PDBeby EMBL-EBI,WellcomeTrust[104948],BBSRC[BB/J007471/1,BB/ K016970/1,BB/K020013/1,BB/M013146/1,BB/M011674/1,BB/M020347/1, BB/M020428/1],NIGMS[1RO1GM079429-01A1],EU[284209,675858] andMRC[MR/L007835/1];PDBjbyJST-NBDCandBMRBbyNIGMS [1R01GM109046].
References
and
recommended
reading
Papers of particular interest, published within the period of review, havebeenhighlightedas:
ofspecialinterest ofoutstandinginterest
1. BernalJD,CrowfootDM:X-rayphotographsofcrystalline pepsin.Nature1934,133:794-795.
2. KendrewJC,BodoG,DintzisHM,ParrishRG,WyckoffH, PhillipsDC:Athree-dimensionalmodelofthemyoglobin moleculeobtainedbyX-rayanalysis.Nature1958,181:662-666.
3. KendrewJC,DickersonRE,StrandbergBE,HartRG,DaviesDR, PhillipsDC,ShoreVC:Structureofmyoglobin:a three-dimensionalFouriersynthesisat2Aresolution.Nature1960,
185:422-427.
4. PerutzMF,RossmannMG,CullisAF,MuirheadH,WillG, NorthACT:Structureofhaemoglobin:athree-dimensional Fouriersynthesisat5.5A˚ resolution,obtainedbyX-ray analysis.Nature1960,185:416-422.
5. BoltonW,PerutzMF:Threedimensionalfouriersynthesisof horsedeoxyhaemoglobinat2.8A˚ ngstromunitsresolution. Nature1970,228:551-552.
6.
ColdSpringHarborSymposiaonQuantitativeBiology.Cold SpringLaboratoryPress;1972.
This seminal meeting highlighted the key structures that had been determinedandbroughttogethertheleadingfiguresinstructuralbiology. ToquoteDavidPhillipsitwasa‘Comingofage.’
7. ProteinDataBank:ProteinDataBank.NatureNewBiol.1971,
233:223.
8. Meyer EFJr,MorimotoCN,VillarrealJ,BermanHM,CarrellHL, StodolaRK,KoetzleTF,AndrewsLC,BernsteinFC,BernsteinHJ etal.:CRYSNET,acrystallographiccomputingnetworkwith interactivegraphicsdisplay.FedProc1974,33:2402-2405.
9. InternationalUnionofCrystallography:Policyonpublicationand thedepositionofdatafromcrystallographicstudiesof biologicalmacromolecules.ActaCryst1989,A45:658.
10.
ReadRJ,AdamsPD,Arendall WB3rd,BrungerAT,EmsleyP, JoostenRP,KleywegtGJ,KrissinelEB,LuttekeT,OtwinowskiZ etal.:Anewgenerationofcrystallographicvalidationtoolsfor theproteindatabank.Structure2011,19:1395-1412.
Thispapercontainedathroughanalysisofmethodsthatcouldbeusedfor validationofstructuresdeterminedbyX-raycrystallography.The recom-mendationswereusedtocreatethevalidationtoolsusedbythewwPDB. 11.
HendersonR,SaliA,BakerML,CarragherB,DevkotaB, DowningKH,EgelmanEH,FengZ,FrankJ,GrigorieffNetal.:
Outcomeofthefirstelectronmicroscopyvalidationtaskforce meeting.Structure2012,20:205-214.
Thispaperwasthefirstonetoanalyzewhatisneededtovalidate3DEM mapsandmodels.Itisthebasisforcurrentresearchinthisfield. 12.
MontelioneGT,NilgesM,BaxA,GuntertP,HerrmannT, RichardsonJS,SchwietersCD,VrankenWF,VuisterGW, WishartDSetal.:RecommendationsofthewwPDBNMR validationtaskforce.Structure2013,21:1563-1570.
This paper made recommendations for how to validate structures deter-mined by NMR, and is the basis for current ongoing research. 13.
AdamsPD,AertgeertsK,BauerC,BellJA,BermanHM,BhatTN, BlaneyJM,BoltonE,BricogneG,BrownDetal.:Outcomeofthe firstwwPDB/CCDC/D3Rligandvalidationworkshop.Structure 2016,24:502-508.
The criteria for judging the quality of ligands in protein complexes are laid outandwillformthebasisforimprovedvalidationofthesemolecules.
14.
BermanHM,HenrickK,NakamuraH:Announcingtheworldwide ProteinDataBank.NatStructBiol2003,10:980.
ThisistheformalannouncementforhowtheProteinDataBankwillbe managedbyaninternationalconsortium.
15. BrownA,BernalJD:TheSageofScience. Oxford:Oxford UniversityPress;2005.
16.
LawsonCL,PatwardhanA,BakerML,HrycC,GarciaES, HudsonBP,LagerstedtI,LudtkeSJ,PintilieG,SalaRetal.:
EMDataBankunifieddataresourcefor3DEM.NucleicAcids Res2016,44:D396-D403.
Thispaperdescribestheproceduresforstreamliningthedepositionand distributionof3DEMmapsandmodels.
17. IudinA,KorirPK,Salavert-TorresJ,KleywegtGJ,PatwardhanA:
EMPIAR:apublicarchiveforrawelectronmicroscopyimage data.NatMethods2016:13.
18.
BermanHM,BurleySK,ChiuW,SaliA,AdzhubeiA,BournePE, BryantSH,DunbrackRLJr,FidelisK,FrankJetal.:Outcomeofa workshoponarchivingstructuralmodelsofbiological macromolecules.Structure2006,14:1211-1217.
TherecommendationtoremovepurelyinsilicomodelsfromthePDBis contained in this paper.
19. HaasJ,RothS,ArnoldK,KieferF,SchmidtT,BordoliL, SchwedeT:TheProteinModelPortal–acomprehensive resourceforproteinstructureandmodelinformation. Database(Oxford)2013,2013:bat031.
20.
FitzgeraldPMD,WestbrookJD,BournePE,McMahonB, WatenpaughKD,BermanHM:4.5Macromoleculardictionary (mmCIF).In InternationalTablesforCrystallographyG.Definition andExchangeofCrystallographicData.EditedbyHallSR, McMahonB.Springer;2005:295-443.
AcompletedescriptionofthemmCIFdatadictionaryiscontainedhere. 21. MalfoisM,SvergunDI:sasCIF:AnExtensionofCore
CrystallographicInformationFileforSAS.JournalofApplied Crystallography2000,33:812-816.
22. WestbrookJ,ItoN,NakamuraH,HenrickK,BermanHM:PDBML: therepresentationofarchivalmacromolecularstructuredata inXML.Bioinformatics2005,21:988-992.
23.
KinjoAR,SuzukiH,YamashitaR,IkegawaY,KudouT,IgarashiR, KengakuY,ChoH,StandleyDM,NakagawaAetal.:ProteinData BankJapan(PDBj):maintainingastructuraldataarchiveand resourcedescriptionframeworkformat.NucleicAcidsRes 2012,40:D453-D460.
Descriptions of PDBj services are given here as well as the RDF format. 24. LinD,ManningNO,JiangJ,AbolaEE,StampfD,PriluskyJ,
SussmanJL:AutoDep:aweb-basedsystemfordepositionand validationofmacromolecularstructuralinformation.Acta Cryst2000,D56:828-841.
25. TagariM,TateJ,SwaminathanGJ,NewmanR,NaimA, VrankenW,KapopoulouA,HussainA,FillonJ,HenrickKetal.: E-MSD:improvingdatadepositionandstructurequality.Nucleic AcidsRes2006,34:D287-D290.
26.
BermanHM,WestbrookJ,FengZ,GillilandG,BhatTN,WeissigH, ShindyalovIN,BournePE:TheProteinDataBank.NucleicAcids Res2000,28:235-242.
Thisisthefirstcompletedescriptionoftheservicesprovided bythe RCSBPDB
27.
UlrichEL,AkutsuH,DoreleijersJF,HaranoY,IoannidisYE,LinJ, LivnyM,MadingS,MaziukD,MillerZetal.:BioMagResBank. NucleicAcidsRes2008,36:D402-D408.
A summary of the services provided by BMRB is given here. 28. DrugDesignDataResourceCommunity:ContinuousEvaluationof
LigandPosePrediction.https://drugdesigndata.org/about/celpp
(accessed18.04.16).
29. RosePW,PrlicA,BiC,BluhmWF,ChristieCH,DuttaS,GreenRK, GoodsellDS,WestbrookJD,WooJetal.:TheRCSBProteinData Bank:viewsofstructuralbiologyforbasicandapplied researchandeducation.NucleicAcidsRes2015,43:D345-D356.
30. VelankarS,vanGinkelG,AlhroubY,BattleGM,BerrisfordJM, ConroyMJ,DanaJM,GoreSP,GutmanasA,HaslamPetal.:
datafromPDBandEMDB.NucleicAcidsRes2016,44: D385-D395.
31.
GutmanasA,AlhroubY,BattleGM,BerrisfordJM,BochetE, ConroyMJ,DanaJM,FernandezMonteceloMA,vanGinkelG, GoreSPetal.:PDBe:ProteinDataBankinEurope.Nucleic AcidsRes2014,42:D285-D291.
The services offered by PDBe are described.
32. KinjoAR,NakamuraH:Compositestructuralmotifsofbinding sitesfordelineatingbiologicalfunctionsofproteins.PLoSOne 2012,7:e31437.
33. MarkleyJL,UlrichEL,BermanHM,HenrickK,NakamuraH, AkutsuH:BioMagResBank(BMRB)asapartnerinthe WorldwideProteinDataBank(wwPDB):newpolicies affectingbiomolecularNMRdepositions.JBiomolNMR2008,
40:153-155.
34. SillitoeI,LewisTE,CuffA,DasS,AshfordP,DawsonNL, FurnhamN,LaskowskiRA,LeeD,LeesJGetal.:CATH: comprehensivestructuralandfunctionalannotationsfor genomesequences.NucleicAcidsRes2015,43:D376-D381.
35. AndreevaA,HoworthD,BrennerSE,HubbardTJ,ChothiaC, MurzinAG:SCOPdatabasein2004:refinementsintegrate structureandsequencefamilydata.NucleicAcidsRes2004,
32:D226-D229.
36. AndreevaA,HoworthD,ChandoniaJM,BrennerSE,HubbardTJ, ChothiaC,MurzinAG:DatagrowthanditsimpactontheSCOP database:newdevelopments.NucleicAcidsRes2008,
36:D419-D425.
37. LewisTE,SillitoeI,AndreevaA,BlundellTL,BuchanDW, ChothiaC,CozzettoD,DanaJM,FilippisI,GoughJetal.:
Genome3D:exploitingstructuretohelpusersunderstand theirsequences.NucleicAcidsRes2015,43:D382-D386.
38. BermanHM,OlsonWK,BeveridgeDL,WestbrookJD,GelbinA, DemenyT,HsiehS-h,SrinivasanAR,SchneiderB:TheNucleic AcidDatabase–acomprehensiverelationaldatabaseof three-dimensionalstructuresofnucleicacids.Biophys.J. 1992,63:751-759.
39. GabanyiMJ,AdamsPD,ArnoldK,BordoliL,CarterLG, Flippen-AndersenJ,GiffordL,HaasJ,KouranovA,McLaughlinWAetal.:
TheStructuralBiologyKnowledgebase:aportaltoprotein structures,sequences,functions,andmethods.JStructFunct Genom2011,12:45-54.
40. MadlT,GabelF,SattlerM:NMRandsmall-angle scattering-basedstructuralanalysisofproteincomplexesinsolution.J StructBiol2011,173:472-482.
41. WangZ,ChernyshevA,KoehnEM,ManuelTD,LesleySA, KohenA:Oxidaseactivityofaflavin-dependentthymidylate synthase.FEBSJ2009,276:2801-2810.
42. ByeonIJ,LouisJM,GronenbornAM:Acapturedfolding intermediateinvolvedindimerizationanddomain-swapping ofGB1.JMolBiol2004,340:615-625.
43. WardAB,SaliA,WilsonIA:Biochemistry.Integrativestructural biology.Science2013,339:913-915.
44.
SaliA,BermanHM,SchwedeT,TrewhellaJ,KleywegtG, BurleySK,MarkleyJ,NakamuraH,AdamsP,BonvinAMetal.:
OutcomeofthefirstwwPDBhybrid/integrativemethodstask forceworkshop.Structure2015,23:1156-1167.
This paper summarized the steps necessary to establish an archive of structure data from hybrid/integrative methods.