JOURNAL OFCLINICALMICROBIOLOGY, 0095-1137/97/$04.0010
Jan. 1997, p. 273–277 Vol. 35, No. 1
Copyrightq1997, American Society for Microbiology
Use of a PCR Method Based on IS6110 Polymorphism for
Typing Mycobacterium tuberculosis Strains from
BACTEC Cultures
ISABEL OTAL,* SOFI´A SAMPER, M. PILAR ASENSIO, M. ASUNCIO´ N VITORIA, M. CARMEN RUBIO, RAFAEL GO´ MEZ-LUS,ANDCARLOS MARTI´N
Departamento de Microbiologı´a, Medicina Preventiva, y Salud Pu´blica, Universidad de Zaragoza,
50009 Zaragoza, Spain
Received 25 July 1996/Returned for modification 5 September 1996/Accepted 3 October 1996
Two PCR typing methods, based on polymorphism of the insertion sequence IS6110, were compared with
Mycobacterium tuberculosisstrains by using a single primer complementary to the inverted repeats of IS6110. TotalM. tuberculosisDNA either was amplified directly (IS6110-PCR) or was amplified following digestion and ligation (IS6110-inverse-PCR). Both PCR techniques showed a similar degree of discrimination. Because of its simplicity, IS6110-PCR was chosen to confirm that a singleM. tuberculosis strain was responsible for an outbreak of tuberculosis in a secondary school. IS6110-PCR was used to study the degree of differentiation in 85 clinicalM. tuberculosisisolates from BACTEC 12B broth cultures. Results were consistent with those of the standardized IS6110 restriction fragment length polymorphism (RFLP) analysis method, showing identical PCR types for identical RFLPs, although the degree of discrimination was greater by RFLP analysis. The study concludes that due to its simplicity, IS6110-PCR is a good screening method when quick differentiation between
M. tuberculosisstrains is needed because BACTEC cultures may be used directly.
Epidemiological studies of tuberculosis can be greatly facil-itated by the application of strain-specific markers. In order to type Mycobacterium tuberculosis complex strains, most molec-ular methods are based on the detection of repetitive elements (4, 8). The repetitive elements described so far in M.
tubercu-losis are the following: the insertion sequences IS6110 and
IS1081, the major polymorphic tandem repeats (MPTRs), the polymorphic GC-rich repetitive sequences (PGRSs), and the direct repeat sequence (DR) (7, 14). IS6110 is distributed throughout the M. tuberculosis complex (15). At present the method most extensively used to differentiate M. tuberculosis strains is the internationally standardized (16) Southern blot-ting-based technique, consisting of restriction fragment length polymorphism (RFLP) analysis by probing with the insertion sequence IS6110. Its use allows for the comparison of DNA “fingerprints” of M. tuberculosis strains of different origins and from different laboratories. However, RFLP analysis is a time-consuming method that requires growth of the organism fol-lowed by purification of genomic DNA from bacteria.
The development of new PCR typing methods attempts to decrease the time for the differentiation of M. tuberculosis strains. Three approaches have been used to detect polymor-phisms: first, the use of nonspecific primers; second, the use of primers based on the polymorphism of the IS6110 flanking regions; and third, the use of primers based on the DR regions. The methods based on random amplified polymorphism DNA analysis with nonspecific oligonucleotide primers de-scribed previously (6, 9) indicate that standardization and reproducibility as well as comparison between patterns are difficult.
Most of the PCR-based techniques have been developed in order to amplify polymorphic DNA regions flanking IS6110 by
PCR with oligonucleotide primers to the end of IS6110 (11, 13). Other approaches combining the variability of the IS6110 insertion sites with the conserved sites of the MPTR (12) or PGRS (2) for differentiating M. tuberculosis strains have been described, while methods based on IS6110 polymorphism analysis use one primer specific for IS6110 and a second primer complementary to a linker ligated to the restricted genomic DNA (1, 5, 10). For most of these methods sequen-tial DNA manipulations such as digestion, ligation, and amplification are necessary, which complicate their use. A novel, promising method of strain differentiation has been developed. The method is based on the nature of the DNA polymorphism in the spacer regions between the DR and is called spoligotyping. This method allows for the rapid detec-tion and simultaneous differentiadetec-tion of M. tuberculosis strains (3, 17).
Here we describe the use of two PCR typing methods am-plifying the flanking sequences on both sides of IS6110, IS6110-PCR and IS6110-inverse-PCR, and we compare the results with the well-standardized RFLP typing method. We also evaluate the ability of the IS6110-PCR method to detect outbreaks and to differentiate M. tuberculosis strains from BACTEC 12 B broth cultures. Some of the advantages of PCR techniques are their simplicity and speed, while the use of BACTEC 12 B cultures means that results can be obtained in 4 days, not the 4 to 5 weeks required to obtain growth on solid media. IS6110-PCR is a simplified method based on that scribed by Ross and Dwyer (13), but the IS6110-PCR de-scribed here uses only one primer rather than the two primers used by Ross and Dwyer (13). The oligonucleotide primer designed was complementary to the inverted repeat of IS6110 (Fig. 1). The 39 end of the primer is directed outward from both sides of the element. The location of PCR primer CAR-2 in the insertion sequence is shown in Fig. 1. DNA samples were denatured by incubation for 3 min at 948C before amplification for 40 cycles of 948C for 1 min, 628C for 1 min, and 728C for 2 min. The reaction was performed in a Perkin-Elmer Cetus
* Corresponding author. Mailing address: Departamento de Micro-biologı´a, Medicina Preventiva, y Salud Pu´blica, Universida de Zara-goza, Domingo Miral s/n, 50009 ZaraZara-goza, Spain. Phone: 34-76-761759. Fax: 34-76-761693. E-mail: otali@posta.unizar.es.
273
on May 15, 2020 by guest
http://jcm.asm.org/
DNA thermal cycler. PCR products were analyzed by electro-phoresis in 1.4% agarose gels or 8% acrylamide gels and were stained with ethidium bromide or silver stain, respectively. IS6110-inverPCR was used to amplify the flanking se-quences on both sides of the IS because more bands were expected for IS6110-inverse-PCR than for IS6110-PCR. IS6110-inverse-PCR is based on PstI digestion of total DNA (no PstI site is present in IS6110) and PCR after ligation (Fig. 1). DNA samples were digested with PstI. The enzyme was inactivated for 10 min at 708C. Ligation was performed at 168C for 4 h. Amplification of 1 ml of the ligation mixture was performed as described before for IS6110-PCR.
To determine the discriminating power of the PCR methods to distinguish between epidemiologically unrelated isolates, we examined nine randomly chosen strains which were previously shown to be different by RFLP analysis and we carried out IS6110-PCR, IS6110-inverse-PCR, and IS6110-RFLP analysis. As shown in Fig. 2, all strains exhibited different banding pat-terns by the IS6110-RFLP method, and less resolution was observed by the IS6110-PCR method (Fig. 2B). Isolates 7 and 9 showed the same pattern as isolates 6, 7, and 9 by IS6110-inverse-PCR (Fig. 2C). To determine if strains which gave identical RFLP patterns were identical by PCR, we chose the simpler and quicker method of PCR, IS6110-PCR (digestion
and ligation are not needed) to analyze two groups of four isolates from our hospital (HCU, Zaragoza, Spain) which had previously been shown to have the same RFLP pattern. The pattern obtained by IS6110-PCR was also found to be the same for the isolates of each group (Fig. 3).
[image:2.612.69.547.61.403.2]In order to demonstrate the usefulness of the IS6110-PCR in the detection of outbreaks, we selected a further six isolates from an outbreak in a secondary school. All patients were students and classmates whose isolates had the same RFLP pattern, and we found the same IS6110-PCR pattern (Fig. 4). In order to ensure that the IS6110-PCR method is reproduc-ible, we worked with different concentrations of DNA from one sample. We then obtained dilutions containing concentra-tions from 50 ng/ml to 0.5 pg/ml, and the IS6110-PCR patterns obtained were the same. Likewise, nine M. tuberculosis strains of different origins that were sent from the Institute for Public Health and Hygiene in Bilthoven, The Netherlands, as part of a reproducibility project were analyzed by the RFLP method and IS6110-PCR. Nine different patterns were obtained by the RFLP method, while only seven patterns were obtained by the IS6110-PCR method (data not shown). Although there is less discrimination between strains, this PCR method is useful for obtaining preliminary data and can be performed with FIG. 1. Location of PCR primer CAR2 (GACIIICCGGGGCGGTTCA) in the insertion sequence IS6110. The oligonucleotide primer designed was described previously (4) and is present at the inverted repeat of IS6110. The 39end of the primer is directed outward from both sides of the element. (A) IS6110-PCR typing.
(B) IS6110-inverse-PCR.
on May 15, 2020 by guest
http://jcm.asm.org/
BACTEC cultures without having to wait for growth on solid media, as is the case for RFLP analysis.
The ability of the IS6110-PCR method to differentiate M.
tuberculosis isolates from BACTEC 12 B broth cultures was
evaluated by analyzing 85 samples. In preparation for PCR, 300 ml from the BACTEC 12 B vials with a growth index of
.150 was heated at 858C for 10 min, and the sample was pelleted by centrifugation at 8,0003g for 5 min. The
super-natant was flicked off, and the pellet was emulsified in 50ml of chloroform. Distilled water was added to bring the total vol-ume of the aqueous phase to 50 ml, and this was emulsified with the chloroform phase. After centrifugation at 8,0003 g
for 3 min, 5ml of the aqueous supernatant was used in a total volume of 50ml of the PCR mixture. The samples were stored at2208C without removing the chloroform and were recentri-fuged prior to use. Amplification was performed as described before for IS6110-PCR. We analyzed 85 routine isolates of M.
tuberculosis from our hospital from April 1994 to October 1995
by PCR in BACTEC 12 B medium. Two M. tuberculosis strains with two copies of IS6110 were eliminated from the study because the discriminatory power of RFLP analysis is known to be decreased with a low copy number of IS6110 and since our results were being compared with the corresponding RFLPs detected by hybridization with IS6110. The PCR and RFLP patterns were scanned, and the corresponding dendrograms were obtained by using the GelCompare program (Applied-Maths, Kortrijk, Belgium). When the samples were different by PCR, we always observed different RFLP patterns. When some samples were identical by RFLP analysis, the pattern obtained by PCR was the same. Identity by IS6110-PCR, how-ever, requires further typing investigations, because due to its limited resolution, identity by IS6110-PCR does not necessarily imply strain identity. The pattern of amplicons produced could be used to cluster strains of M. tuberculosis into groups which correlated with the groups formed by typing by IS6110-RFLP analysis (Fig. 5). We obtained 47 different patterns by PCR and 72 by RFLP analysis.
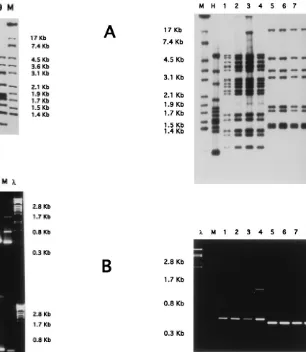
[image:3.612.227.533.68.420.2]The possibility of using PCR with cultures from BACTEC 12 B vials is of particular interest for use as a rapid response to FIG. 2. Nine randomly chosen strains which were previously shown to be
different by RFLP were compared. (A) RFLP Southern blot hybridization with IS6110. (B) IS6110-PCR. (C) IS6110-inverse-PCR in a 1.4% agarose gel. Lanes: M, control strain M. tuberculosis 14323; 1 to 9, clinical isolates;l, bacteriophage lDNA digested with PstI.
FIG. 3. Two sets of four different isolates showing identical banding patterns. The strains in lanes 1 to 4 are identical and those in lanes 5 to 8 are identical by Southern blot hybridization analysis (A) and IS6110-PCR (B). Lanes: M, control strain M. tuberculosis 14323; H, control strain M. tuberculosis H37Rv; 1 to 8, clinical isolates;l, bacteriophagelDNA digested with PstI.
VOL. 35, 1997 NOTES 275
on May 15, 2020 by guest
http://jcm.asm.org/
[image:3.612.80.344.68.450.2]outbreak situations. Different techniques have been developed to type M. tuberculosis strains, but to our knowledge, this is the first time that the systematic differentiation of M. tuberculosis strains directly from BACTEC 12 B cultures has been re-ported. Our results demonstrate that using IS6110-PCR to differentiate M. tuberculosis strains from BACTEC 12 B cul-tures is a quick and simple method. The size and number of products obtained are limited, but strain identity can be ruled out in 3 to 4 days, compared with the 4 to 5 weeks required for typing by IS6110-RFLP analysis. Therefore, the IS6110-PCR method can be very useful for providing rapid information in outbreak situations and ruling out contamination in the clinical laboratory. It should be emphasized that identity by IS6110-PCR requires further typing investigations, because due to its limited resolution, identity by IS6110-PCR does not necessarily imply strain identity. The PCR typing method from BACTEC 12 B cultures may be used as a quick means of indirectly detecting M. tuberculosis multidrug-resistant strains.
This work was supported by grant FIS/0051 from the Spanish Fondo de Investigaciones Sanitarias de la Seguridad Social, by the European Commission CA on the Epidemiology on Tuberculosis BIOMED1, and grants CT93-1614 and BIO95-0675 from CICYT. Sofı´a Samper was the recipient of a FIS fellowship.
We thank Julie-Ann Gavigan for critical reading of the manuscript, Lorena Lo´pez and Alfonso Barbera for help in DNA extractions, and Jesu´s Molina for supplying samples.
FIG. 4. Six different isolates from an outbreak showing identical banding patterns by Southern blot hybridization analysis (A) and IS6110-PCR (B). Lanes: 1 to 6, clinical isolates; M, control strain M. tuberculosis 14323.
FIG. 5. Dendrogram based on computer-assisted comparison of patterns ob-tained by RFLP (R-1 to R-72) correlated with the patterns obob-tained by IS6110-PCR (P-1 to P-47).
276
on May 15, 2020 by guest
REFERENCES
1. Butler, W. R., W. H. Haas, and J. T. Crawford. 1996. Automated DNA fingerprinting analysis of Mycobacterium tuberculosis using fluorescent de-tection of PCR products. J. Clin. Microbiol. 34:1801–1803.
2. Friedman, C. R., M. Y. Stoeckle, W. D. Johnson, Jr., and L. W. Riley. 1995. Double-repetitive-element PCR method for subtyping Mycobacterium tuber-culosis clinical isolates. J. Clin. Microbiol. 33:1383–1384.
3. Groenen, P. M. A., A. E. Bunschoten, D. van Soolingen, and J. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Myco-bacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 10:1057–1065.
4. Gutierrez, M., S. Samper, J. A. Gavigan, J. F. Garcı´a Marı´n, and C. Martı´n. 1995. Differentiation by molecular typing Mycobacterium bovis strains caus-ing tuberculosis in cattle and goats. J. Clin. Microbiol. 33:2953–2956. 5. Haas, W. H., W. R. Butler, C. L. Wooley, and J. T. Crawford. 1993.
Mixed-linker polymerase chain reaction: a new method for rapid fingerprinting of isolates of the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 31: 1293–1298.
6. Linton, C. J., H. Jalal, J. P. Leeming, and M. R. Millar. 1994. Rapid discrimination of Mycobacterium tuberculosis strain by random amplified polymorphic DNA analysis. J. Clin. Microbiol. 32:2169–2174.
7. Martı´n, C., S. Samper, I. Otal, P. Asensio, R. Gomez-Lus, G. Torrea, and B.
Gicquel.1994. New methods for diagnosis and epidemiological studies of tuberculosis based on PCR and RFLP, p. 105–113. In F. G. Priest (ed.), Bacterial diversity and systematics. Plenum, New York, N.Y.
8. Otal, I., C. Martı´n, V. Vicent-Le´vy-Fre´bault, D. Thierry, and B. Gicquel. 1991. Restriction fragment length polymorphism analysis using IS6110 as epidemiological marker in tuberculosis. J. Clin. Microbiol. 29:1252–1254. 9. Palittapongarnpim, P., S. Chomyc, A. Fanning, and D. Kunimoto. 1993.
DNA fragment length polymorphism analysis of Mycobacterium tuberculosis
isolates by arbitrarily primed polymerase chain reaction. J. Infect. Dis. 167: 975–978.
10. Palittapongarnpim, P., S. Chomyc, A. Fanning, and D. Kunimoto. 1993. DNA fingerprinting of Mycobacterium tuberculosis isolates by ligation-medi-ated polymerase chain reaction. Nucleic Acids Res. 21:761–762.
11. Patel, S., S. Wall, and N. A. Saunders. Heminested inverse PCR for IS6110 fingerprinting of Mycobacterium tuberculosis strains. J. Clin. Microbiol. 34: 1686–1690.
12. Plikaytis, B. B., J. T. Crawford, C. L. Woodley, W. R. Butler, K. D. Eisenach,
M. D. Cave, and T. M. Shinnick.1993. Rapid, amplification-based finger-printing of Mycobacterium tuberculosis. J. Gen. Microbiol. 139:1537–1542. 13. Ross, B., and B. Dwyer. 1993. Rapid, simple method for typing isolates of
Mycobacterium tuberculosis by using the polymerase chain reaction. J. Clin. Microbiol. 31:329–334.
14. Small, P. M., and J. D. A. van Embden. 1994. Molecular epidemiology of tuberculosis, p. 569–582. In B. Bloom (ed.), Tuberculosis. ASM Press, Wash-ington, D.C.
15. Thierry, D., A. Brisson-Noel, V. Vincent-Le´vy-Fre´bault, S. Nguyen, J. L.
Guesdon, and B. Gicquel.1990. Characterization of a Mycobacterium tuber-culosis insertion sequence, IS6110, and its application in diagnosis. J. Clin. Microbiol. 28:2668–2673.
16. Van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D.
Eisenach, B. Gicquel, P. Hermans, C. Martı´n, R. McAdam, T. Shinnick, and P. Small.1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406–409.
17. Van Soolingen, D., L. Quian, P. E. De Haas, J. T. Douglas, H. Traatore, F.
Portaels, H. Z. Quing, D. Enkhsaikan, P. Nymadawa, and J. D. A. Van Embden.1995. Predominance of a single genotype of Mycobacterium tuber-culosis in countries of East Asia. J. Clin. Microbiol. 33:3234–3238.
VOL. 35, 1997 NOTES 277