organic papers
o2462
Yanet al. C30H25N5O4 doi:10.1107/S160053680601854X Acta Cryst.(2006). E62, o2462–o2463
Acta Crystallographica Section E
Structure Reports
Online
ISSN 1600-5368
1-Acetyl-3,3-bis[3-(4-methylphenyl)-1,2,4-oxadiazol-5-ylmethyl]-1
H
-indol-2(3
H
)-one
Xiao-Chen Yan,* Hai-Bo Wang and Zhi-Qian Liu
Department of Applied Chemistry, College of Science, Nanjing University of Technology, Xinmofan Road No. 5 Nanjing, Nanjing 210009, People’s Republic of China
Correspondence e-mail: wanghaibo@njut.edu.cn
Key indicators
Single-crystal X-ray study T= 293 K
Mean(C–C) = 0.006 A˚ Rfactor = 0.043 wRfactor = 0.127 Data-to-parameter ratio = 8.1
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
Received 15 May 2006 Accepted 18 May 2006
#2006 International Union of Crystallography All rights reserved
In the crystal structure of the title compound, C30H25N5O4,
there are C—H O and C—H N hydrogen bonds.
Comment
Oxindole derivatives, which possess affinity for different receptors, exhibit intrinsic analgesic (Daisley & Walker, 1979), anti-inflammatory (Kadin, 1986), antiviral (Singh & Krishna, 1989), cardiotonic (Andreani & Rambaldi, 1988), anti-convulsant (Valentaet al., 1990), anxiolytic (Sargeset al., 1989) and inotropic (Ogawaet al., 1988) properties. We are focusing our synthetic and structural studies on new oxindole deriva-tives, and recently we have published the synthesis and structure of 1-acetyl-3,3-bis[(3-(2-methylphenyl)-1,2,4-oxa-diazol-5-yl)methyl]-1H-indolin-2(3H)-one (Yan et al., 2006). We report here the structure of its close analogue, (I), with 2-methylphenyl replaced by the 4-methylphenyl group.
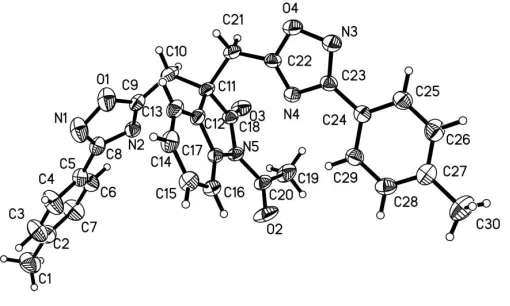
The molecular structure of (I) is shown in Fig. 1. The indanone ring system is planar and the acetyl group at N5 is twisted by 17.5 (2). The dihedral angle between the N4/ C22O4/N3/C23 and C24–C29 planes is 11.1 (1) and that between the N2/C8/N1/O1/C29 and C2–C7 planes is 25.6 (1). Intramolecular C—H N and C—H O hydrogen bonds are observed in the molecular structure. The crystal structure is stabilized by intermolecular C—H O and C—H N hydrogen bonds (Table 1).
Experimental
was obtained. The pure compound was obtained by crystallization from a mixture of ethyl acetate (4 ml) and petrolum ether (8 ml). Crystals of (I) suitable for X-ray diffraction were obtained by slow evaporation of an ethanol solution.
Crystal data
C30H25N5O4
Mr= 519.55
Monoclinic,C2
a= 15.5510 (16) A˚
b= 6.9695 (14) A˚
c= 25.220 (2) A˚
= 103.46 (3)
V= 2658.3 (7) A˚3
Z= 4
Dx= 1.298 Mg m
3
MoKradiation
= 0.09 mm1
T= 293 (2) K Block, colourless 0.400.400.30 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
!/2scans
Absorption correction: scan (Northet al., 1968)
Tmin= 0.965,Tmax= 0.974
5676 measured reflections
2843 independent reflections 2200 reflections withI> 2(I)
Rint= 0.030 max= 26.0
3 standard reflections every 200 reflections intensity decay: none
Refinement
Refinement onF2
R[F2> 2(F2)] = 0.044
wR(F2) = 0.127
S= 1.05 2843 reflections 353 parameters
H-atom parameters constrained
w= 1/[2(F
o2) + (0.075P)2
+ 0.19P]
whereP= (Fo2+ 2Fc2)/3
(/)max< 0.001 max= 0.16 e A˚
3
min=0.15 e A˚ 3
Extinction correction:SHELXL97
Extinction coefficient: 0.0098 (12)
Table 1
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
C7—H7A N1i 0.93 2.62 3.387 (6) 140 C10—H10A O2ii
0.97 2.46 3.411 (5) 166 C16—H16A O2 0.93 2.30 2.834 (5) 116 C29—H29A N4 0.93 2.62 2.935 (4) 100
Symmetry codes: (i)x;y1;z; (ii)x1 2;yþ
1 2;z.
All H atoms were positioned geometrically with C—H distances in the range 0.93–0.97 A˚ and included in the refinement in the riding-model approximation, withUiso(H) values of 1.2 or 1.5 timesUeq(C).
In the absence of significant anomalous scattering, Friedel pairs were merged.
Data collection: CAD-4 Software (Enraf–Nonius, 1989); cell refinement: CAD-4 Software; data reduction: XCAD4 (Harms & Wocadlo, 1995); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure:SHELXL97 (Sheldrick, 1997); molecular graphics:SHELXTL (Siemens, 1996); software used to prepare material for publication:SHELXL97.
References
Andreani, A. & Rambaldi, M. (1988).J. Heterocycl. Chem.25, 1519–1523. Daisley, R. W. & Walker, J. (1979).Eur. J. Med. Chem. Chim. Ther.14, 47–52. Enraf–Nonius (1989).CAD-4 Software. Version 5.0. Enraf–Nonius, Delft, The
Netherlands.
Harms, K. & Wocadlo, S. (1995).XCAD4. University of Marburg, Germany. Kadin, S. B. (1986). Eur. Patent No. EP175551.
North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968).Acta Cryst.A24, 351– 359.
Ogawa, H., Tamada, S., Fujioka, T., Teramoto, S., Kondo, K., Yamashita, S., Yabuuchi, Y., Tominaga, M. & Nakagawa, K. (1988).Chem. Pharm. Bull.36, 2253–2258.
Sarges, R., Howard, H. R., Koe, B. K. & Weissman, A. (1989).J. Med. Chem.
32, 437–444.
Sheldrick, G. M. (1997). SHELXL97 and SHELXS97. University of Go¨ttingen, Germany.
Siemens (1996). SHELXTL. Version 5.06. Siemens Analytical X-ray Instruments Inc., Madison, Wisconsin, USA.
Singh, S. P. & Krishna, J. (1989).Zentralbl. Mikrobiol.144, 105–109. Valenta, V., Holubek, J., Svatek, E., Valchar, M., Krejci, I. & Protiva, M.
(1990).Collect. Czech. Chem. Commun.55, 2756–2764.
[image:2.610.315.570.71.224.2]Yan, X.-C., Wang, H.-B. & Liu, Z.-Q. (2006).Acta Cryst.E62, o917–o918.
Figure 1
supporting information
sup-1
Acta Cryst. (2006). E62, o2462–o2463
supporting information
Acta Cryst. (2006). E62, o2462–o2463 [https://doi.org/10.1107/S160053680601854X]
1-Acetyl-3,3-bis[3-(4-methylphenyl)-1,2,4-oxadiazol-5-ylmethyl]-1
H
-indol-2(3
H
)-one
Xiao-Chen Yan, Hai-Bo Wang and Zhi-Qian Liu
1-Acetyl-3,3-bis[3-(4-methylphenyl)-1,2,4-oxadiazol-5-ylmethyl]-1H- indol-2(3H)-one
Crystal data
C30H25N5O4
Mr = 519.55
Monoclinic, C2 Hall symbol: C 2y
a = 15.5510 (16) Å
b = 6.9695 (14) Å
c = 25.220 (2) Å
β = 103.46 (3)°
V = 2658.3 (7) Å3
Z = 4
F(000) = 1088
Dx = 1.298 Mg m−3
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 25 reflections
θ = 10–13°
µ = 0.09 mm−1
T = 293 K Block, colourless 0.40 × 0.40 × 0.30 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
ω/2θ scans
Absorption correction: ψ scan (North et al., 1968)
Tmin = 0.965, Tmax = 0.974
5676 measured reflections
2843 independent reflections 2200 reflections with I > 2σ(I)
Rint = 0.030
θmax = 26.0°, θmin = 1.7°
h = −19→19
k = 0→8
l = −31→31
3 standard reflections every 200 reflections intensity decay: none
Refinement
Refinement on F2
Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.044
wR(F2) = 0.127
S = 1.05 2843 reflections 353 parameters 1 restraint
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained
w = 1/[σ2(F
o2) + (0.075P)2 + 0.19P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.16 e Å−3
Δρmin = −0.15 e Å−3
Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4
Special details
Experimental. 1H NMR (CDCl
3, δ, p.p.m.): 8.16 (m, 1H), 7.74–7.75 (s, 4H), 7.19–7.23 (s, 5H), 7.03–7.05 (s, 2H), 3.74–
3.77 (m, 2H), 3.63–3.66 (m, 2H), 2.75 (s, 3H), 2.35 (s, 6H).
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
supporting information
sup-3
Acta Cryst. (2006). E62, o2462–o2463
H14A 0.6435 1.1888 0.8350 0.072* C15 0.7261 (2) 0.9816 (6) 0.82098 (16) 0.0593 (9) H15A 0.7779 1.0444 0.8383 0.071* C16 0.7320 (2) 0.8019 (5) 0.79763 (14) 0.0493 (8) H16A 0.7865 0.7445 0.7990 0.059* C17 0.65412 (18) 0.7129 (5) 0.77239 (12) 0.0406 (7) C18 0.54967 (19) 0.4971 (5) 0.72633 (13) 0.0448 (8) C19 0.6825 (3) 0.2680 (7) 0.68912 (18) 0.0757 (13) H19A 0.7348 0.2108 0.6819 0.114* H19B 0.6460 0.3162 0.6557 0.114* H19C 0.6503 0.1733 0.7043 0.114* C20 0.7080 (2) 0.4284 (6) 0.72852 (15) 0.0531 (9) C21 0.4433 (2) 0.7608 (6) 0.68741 (14) 0.0531 (9) H21A 0.4003 0.6666 0.6695 0.064* H21B 0.4108 0.8693 0.6969 0.064* C22 0.4952 (2) 0.8266 (6) 0.64798 (13) 0.0488 (8) C23 0.5851 (2) 0.8925 (5) 0.60135 (12) 0.0465 (8) C24 0.6695 (2) 0.9026 (5) 0.58370 (13) 0.0454 (7) C25 0.6728 (2) 0.9900 (6) 0.53497 (15) 0.0600 (10) H25A 0.6213 1.0362 0.5120 0.072* C26 0.7531 (3) 1.0089 (7) 0.52035 (17) 0.0694 (11) H26A 0.7546 1.0681 0.4875 0.083* C27 0.8303 (2) 0.9420 (5) 0.55329 (17) 0.0594 (10) C28 0.8259 (2) 0.8507 (6) 0.60107 (16) 0.0605 (10) H28A 0.8773 0.8013 0.6234 0.073* C29 0.7462 (2) 0.8308 (6) 0.61666 (14) 0.0541 (9) H29A 0.7447 0.7693 0.6492 0.065* C30 0.9181 (3) 0.9625 (8) 0.5372 (2) 0.0872 (15) H30A 0.9643 0.9085 0.5652 0.131* H30B 0.9300 1.0959 0.5328 0.131* H30C 0.9153 0.8960 0.5035 0.131*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
C5 0.061 (2) 0.047 (2) 0.058 (2) −0.0034 (18) 0.0207 (17) −0.0037 (18) C6 0.080 (3) 0.051 (2) 0.071 (3) 0.002 (2) 0.003 (2) −0.010 (2) C7 0.111 (4) 0.046 (3) 0.091 (4) 0.008 (3) 0.002 (3) 0.003 (3) C8 0.060 (2) 0.044 (2) 0.063 (2) −0.0014 (18) 0.0252 (17) −0.0040 (18) C9 0.0471 (17) 0.049 (2) 0.065 (2) 0.0032 (17) 0.0303 (16) 0.0028 (19) C10 0.0386 (16) 0.057 (2) 0.075 (2) 0.0088 (17) 0.0218 (15) 0.005 (2) C11 0.0347 (14) 0.0447 (19) 0.0560 (18) 0.0113 (15) 0.0160 (13) 0.0038 (16) C12 0.0427 (16) 0.0440 (19) 0.0504 (17) 0.0105 (15) 0.0200 (13) 0.0024 (16) C13 0.0532 (18) 0.046 (2) 0.066 (2) 0.0104 (18) 0.0269 (16) 0.0005 (19) C14 0.075 (2) 0.043 (2) 0.070 (2) 0.0036 (19) 0.0296 (19) −0.0085 (19) C15 0.057 (2) 0.051 (2) 0.070 (2) −0.0055 (19) 0.0159 (17) −0.010 (2) C16 0.0412 (16) 0.046 (2) 0.061 (2) 0.0049 (16) 0.0127 (14) 0.0000 (18) C17 0.0386 (15) 0.0382 (17) 0.0457 (16) 0.0081 (14) 0.0113 (12) 0.0029 (15) C18 0.0373 (16) 0.043 (2) 0.0529 (19) 0.0070 (15) 0.0080 (13) −0.0011 (17) C19 0.061 (2) 0.071 (3) 0.099 (3) 0.013 (2) 0.027 (2) −0.033 (3) C20 0.0435 (17) 0.047 (2) 0.070 (2) 0.0163 (16) 0.0162 (15) −0.0074 (18) C21 0.0389 (15) 0.057 (2) 0.064 (2) 0.0146 (16) 0.0129 (14) 0.0047 (18) C22 0.0441 (17) 0.050 (2) 0.0498 (18) 0.0141 (16) 0.0061 (13) −0.0001 (17) C23 0.0450 (16) 0.0456 (19) 0.0457 (17) 0.0089 (16) 0.0043 (13) −0.0013 (16) C24 0.0490 (16) 0.0389 (18) 0.0472 (17) 0.0026 (15) 0.0089 (13) −0.0056 (15) C25 0.059 (2) 0.057 (2) 0.064 (2) 0.008 (2) 0.0131 (16) 0.012 (2) C26 0.077 (2) 0.056 (2) 0.082 (3) 0.002 (2) 0.032 (2) 0.017 (2) C27 0.061 (2) 0.043 (2) 0.079 (3) −0.0052 (18) 0.0263 (18) −0.008 (2) C28 0.0473 (18) 0.057 (2) 0.073 (2) 0.0078 (18) 0.0059 (16) −0.009 (2) C29 0.0523 (18) 0.054 (2) 0.0542 (19) 0.0057 (18) 0.0091 (15) 0.0007 (18) C30 0.072 (2) 0.070 (3) 0.132 (4) −0.013 (3) 0.051 (3) −0.011 (3)
Geometric parameters (Å, º)
O1—C9 1.318 (5) C11—C21 1.533 (4)
O1—N1 1.426 (4) C11—C18 1.539 (5)
O2—C20 1.205 (4) C12—C13 1.374 (5)
O3—C18 1.202 (4) C12—C17 1.397 (4)
O4—C22 1.336 (4) C13—C14 1.384 (5)
O4—N3 1.422 (4) C13—H13A 0.9300
N1—C8 1.303 (5) C14—C15 1.384 (5)
N2—C9 1.299 (5) C14—H14A 0.9300
N2—C8 1.369 (5) C15—C16 1.396 (6)
N3—C23 1.294 (4) C15—H15A 0.9300
N4—C22 1.281 (4) C16—C17 1.378 (4)
N4—C23 1.371 (4) C16—H16A 0.9300
N5—C18 1.402 (4) C19—C20 1.487 (5)
N5—C20 1.410 (4) C19—H19A 0.9600
N5—C17 1.432 (4) C19—H19B 0.9600
C1—C2 1.519 (7) C19—H19C 0.9600
C1—H1B 0.9600 C21—C22 1.492 (5)
C1—H1C 0.9600 C21—H21A 0.9700
supporting information
sup-5
Acta Cryst. (2006). E62, o2462–o2463
C2—C7 1.366 (7) C23—C24 1.482 (4)
C2—C3 1.377 (7) C24—C29 1.380 (4)
C3—C4 1.362 (7) C24—C25 1.384 (5)
C3—H3B 0.9300 C25—C26 1.387 (5)
C4—C5 1.374 (6) C25—H25A 0.9300
C4—H4B 0.9300 C26—C27 1.373 (6)
C5—C6 1.378 (6) C26—H26A 0.9300
C5—C8 1.467 (5) C27—C28 1.379 (6)
C6—C7 1.377 (6) C27—C30 1.518 (5)
C6—H6A 0.9300 C28—C29 1.391 (5)
C7—H7A 0.9300 C28—H28A 0.9300
C9—C10 1.467 (5) C29—H29A 0.9300
C10—C11 1.553 (4) C30—H30A 0.9600
C10—H10A 0.9700 C30—H30B 0.9600
C10—H10B 0.9700 C30—H30C 0.9600
C11—C12 1.512 (5)
C2—C7—H7A 118.7 N3—C23—N4 114.7 (3) C6—C7—H7A 118.7 N3—C23—C24 122.0 (3) N1—C8—N2 113.9 (4) N4—C23—C24 123.2 (3) N1—C8—C5 121.0 (4) C29—C24—C25 119.2 (3) N2—C8—C5 125.1 (3) C29—C24—C23 120.3 (3) N2—C9—O1 113.2 (3) C25—C24—C23 120.5 (3) N2—C9—C10 130.5 (3) C24—C25—C26 120.0 (3) O1—C9—C10 116.3 (3) C24—C25—H25A 120.0 C9—C10—C11 114.9 (3) C26—C25—H25A 120.0 C9—C10—H10A 108.5 C27—C26—C25 121.4 (4) C11—C10—H10A 108.5 C27—C26—H26A 119.3 C9—C10—H10B 108.5 C25—C26—H26A 119.3 C11—C10—H10B 108.5 C26—C27—C28 118.1 (3) H10A—C10—H10B 107.5 C26—C27—C30 121.4 (4) C12—C11—C21 114.0 (3) C28—C27—C30 120.5 (4) C12—C11—C18 102.2 (2) C27—C28—C29 121.5 (3) C21—C11—C18 109.8 (3) C27—C28—H28A 119.3 C12—C11—C10 115.4 (3) C29—C28—H28A 119.3 C21—C11—C10 106.3 (2) C24—C29—C28 119.7 (3) C18—C11—C10 109.1 (3) C24—C29—H29A 120.1 C13—C12—C17 120.6 (3) C28—C29—H29A 120.1 C13—C12—C11 129.6 (3) C27—C30—H30A 109.5 C17—C12—C11 109.8 (3) C27—C30—H30B 109.5 C12—C13—C14 119.3 (3) H30A—C30—H30B 109.5 C12—C13—H13A 120.3 C27—C30—H30C 109.5 C14—C13—H13A 120.3 H30A—C30—H30C 109.5 C13—C14—C15 119.7 (4) H30B—C30—H30C 109.5 C13—C14—H14A 120.2
supporting information
sup-7
Acta Cryst. (2006). E62, o2462–o2463
C6—C5—C8—N2 −24.5 (6) C18—C11—C21—C22 −59.4 (4) C8—N2—C9—O1 −2.2 (4) C10—C11—C21—C22 −177.3 (3) C8—N2—C9—C10 179.8 (3) C23—N4—C22—O4 −0.8 (4) N1—O1—C9—N2 1.3 (4) C23—N4—C22—C21 −179.5 (4) N1—O1—C9—C10 179.5 (3) N3—O4—C22—N4 0.8 (5) N2—C9—C10—C11 −85.5 (5) N3—O4—C22—C21 179.6 (3) O1—C9—C10—C11 96.6 (4) C11—C21—C22—N4 14.0 (6) C9—C10—C11—C12 −37.6 (4) C11—C21—C22—O4 −164.6 (3) C9—C10—C11—C21 −164.9 (3) O4—N3—C23—N4 −0.2 (5) C9—C10—C11—C18 76.7 (4) O4—N3—C23—C24 −177.1 (3) C21—C11—C12—C13 62.8 (4) C22—N4—C23—N3 0.6 (4) C18—C11—C12—C13 −178.7 (3) C22—N4—C23—C24 177.5 (3) C10—C11—C12—C13 −60.6 (5) N3—C23—C24—C29 167.6 (4) C21—C11—C12—C17 −117.3 (3) N4—C23—C24—C29 −9.1 (5) C18—C11—C12—C17 1.2 (3) N3—C23—C24—C25 −10.1 (6) C10—C11—C12—C17 119.3 (3) N4—C23—C24—C25 173.2 (3) C17—C12—C13—C14 0.4 (5) C29—C24—C25—C26 −1.5 (6) C11—C12—C13—C14 −179.7 (3) C23—C24—C25—C26 176.2 (4) C12—C13—C14—C15 −0.2 (6) C24—C25—C26—C27 0.1 (7) C13—C14—C15—C16 0.0 (6) C25—C26—C27—C28 1.6 (6) C14—C15—C16—C17 −0.1 (5) C25—C26—C27—C30 179.9 (4) C15—C16—C17—C12 0.3 (5) C26—C27—C28—C29 −1.8 (6) C15—C16—C17—N5 178.9 (3) C30—C27—C28—C29 179.8 (4) C13—C12—C17—C16 −0.5 (5) C25—C24—C29—C28 1.3 (6) C11—C12—C17—C16 179.6 (3) C23—C24—C29—C28 −176.4 (4) C13—C12—C17—N5 −179.3 (3) C27—C28—C29—C24 0.4 (6) C11—C12—C17—N5 0.7 (4)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
C7—H7A···N1i 0.93 2.62 3.387 (6) 140
C10—H10A···O2ii 0.97 2.46 3.411 (5) 166
C16—H16A···O2 0.93 2.30 2.834 (5) 116 C29—H29A···N4 0.93 2.62 2.935 (4) 100