Involvement of a Replicative DNA Helicase of Bacteriophage
T4
in
DNA Recombination
Tetsuro Yonesaki
Department of Biology, College of General Education, Osaka University, Osaka 560, Japan Manuscript received May 23, 1994
Accepted for publication July 6, 1994
ABSTRACT
Bacteriophage T4 gene 41 encodes a replicative DNA helicase that is a subunit of the primosome which is essential for lagging-strand DNA synthesis. A mutation, rrh, was generated and selected in the helicase gene on the basis of limited DNA replication that ceases early. The survival of ultraviolet-irradiated phage and the frequency of genetic recombination are reduced by rrh. In addition, rrh diminishes the production of concatemeric DNA. These results strongly suggest that the gene 41 replicative helicase participates in DNA recombination.
D
NA recombination in bacteriophage T4 is carried o u t by its own gene products (for review see MOSIG 1987). Gene 59 is probably involved in T4 DNA recom- bination (YONESAKI 1994) and is also postulated to func- tion in initiating DNA replication at DNA origins (GAUSS et al. 1994). We have purified gp59 (gene 5 9 protein)and characterized some of its properties. Although
attempts to show an effect of gp59 on in vitro single- strand transfer by UvsX protein, UvsY protein and
gp32
(YONESAKI and MINAGAWA 1989; HASHIMOTO a n d YONESAKI1991) were unsuccessful, we found that
gp59
interacts with gp41, a replicative DNA helicase (YONESAKI 1994). The T4 primosome consists of gp41 and gp61 (a DNA primase). It stimulates the movement of a replication fork and primes lagging-strand DNA synthesis (NOSSALand ALBERTS 1983). The gene 41 DNA helicase binds to single-stranded DNA (ssDNA)
,
hydrolyzes ATP or GTP with a n ssDNA requirement, moves 5' + 3' on ssDNA, and unwinds a duplex when encountered on ssDNA.gp59
has a specific affinity for bothgp32
and gp41, rap- idly binds to ssDNA covered with gp32 (YONESAKI 1994), rescues the ssDNAdependent GTP hydrolysis activity of gp41 from inhibition by gp32, and stimulates lagging- strand DNA synthesis(J.
BARRY a n d B. M. ALBERTS, un- published results and cited in YONESAKI 1994). These findings strongly suggest that gp59 promotes the bind- ing of gp41 to ssDNA when this is inhibited bygp32.
To explain the function of gene 59 in DNA recom- bination, we postulated that gene 59contributes to DNA recombination via the function of gene 41 (YONESAKI 1994).
As
described above, gene 41 is essential for DNA replication. If this gene is also involved in DNA recom- bination, it would be possible to isolate a mutant ofgene 41 which is deficient in DNA recombination but not in DNA replication. This paper describes the isolation a n d properties of a mutation in gene 41 which specifically impairs DNA recombination.Genetics 138: 247-252 (October, 1994)
MATERIALS AND METHODS
Plasmids: Plasmid p415 (a gift of B. M. ALBERTS at University of California, San Francisco) carries a 1936bp T4 insert in pBR322 spanning the T4 restriction map between 20.065
(HindIII) and 22.001 ( ClaI). Its only open reading frame in T4 DNA is gene 41. Plasmid pHA153, derived from p415 after mutagenesis with hydroxylamine, has a 1.2-kb insertion in the region flanked by SacI and HindIII(3) sites (Figure 1) and was found to encode an altered gp41 that migrates slightly slower than wild-type gp41 upon SDSpolyacrylamide gel electro- phoresis (data not shown). The region between SacI and HindIII(3) encodes the Gterminal 11 amino acids of gp41. The 1.2-kb insertion was raised spontaneously and might be an IS element. The production of a protein with slightly reduced mobility could have resulted from an insertion in the C-terminal domain fusing gene 41 and an open reading frame in the inserted DNA. Plasmid p415AHlH3 is a derivative of
p415 from which the sequence flanked by HindIII(1) and HindIII(3) in Figure 1 (and therefore almost all of gene 4 1 ) was deleted, only the N-terminal 14 amino acids out of 475 remaining. Chimeric plasmids p153CE, p153CS, p153EH, p153E.3, p153HS and p153SH were constructed by replacing an appropriate fragment of p415 DNA with the corresponding fragment of pHA153 DNA.
Mutagenesis of plasmid DNA and the selection of gene 41
mutants: Using the method of FREES and FREESE (1964), p415 DNA (200 pg/ml) was incubated at 75" for 30-60 min in 125 mM Na,HPO,, 1 M hydroxylamine-HC1 at a final pH 5.5-6.0, and the reaction was terminated by a 50-fold dilution into 50 mM TrisC1 (pH 7.6), 1 M NaCl, 10% (v/v) acetone. The DNA was further diluted 10-fold with distilled water and was used to transform Escherichia coli K12 MHl (sup' hsdR-) cells. For the primary screening, each ampicillin-resistant clone was ex- amined for the ability to support plaque formation by amN81 (gene 41-) phages on EHA agar. From 500 clones, 33 were selected which supported smaller or fewer plaques than on cells harboring p415. For a subsequent selection for clones causing DNA arrest, T4 DNA synthesis was examined after amN81 infection by measuring the incorporation of ['Hlthy- midine into the trichloroacetic acid (TCA)-insoluble fraction according to the method ofYoNEs.m and MINACAWA (1987). All clones but one showed continuous synthesis of DNA at a low rate compared to cells harboring p415.
248
p 4 1 5
p H A l 5 3
p l 5 3 C S
p l 5 3 S H
p 1 5 3 C E
p l 5 3 E S
p l 5 3 E H
p l 5 3 H S
FIGURE I.-Plasmids. The cloned T4 se- quence in p415 is indicated by the open rectangle. The black arrows designated pBR322 indicate the flanking vector se- quences. The coding region of gene 4 I is bordered by vertical bars and an arrow in- dicates the direction of transcription. Re- striction enzyme sites are presented above the figure. The three Hind111 sites are dis- tinguished by serial numbers in the pa- rentheses. The DNA fragments derived from pHA153 are indicated by shaded squares. A 1.2-kb insertion between SncI and HindlII(3) found in pHA153 is not shown. The pattern of T4 DNA synthesis after infection by a m N d l phage is shown on the right: N = normal, A = arrested.
LB broth contains 10 g Bacto tryptone, 5 g Bacto yeast extract and 10 g NaCl per liter adjusted to pH 7.0. M9A is described in YONESAKI and MINAGAWA (1987). EHA T4 agar medium con- tains 13 g Bacto tryptone, 8 g NaCI, 2 g trisodium citrate.2H20, 1.3 g glucose, and either 10 g agar (for hard bottom agar) or 4 g agar (for soft top agar) per liter.
RESULTS
Isolation of a mutation in gene 42 causing DNA arrest:
The initiation of T4 DNA replication proceeds bimodally (LUDER and
Mosrc
1982). DNA initiation initially depends on DNA origins. Later, origin- dependent initiation is replaced by an alternative mode of initiation that depends on recombination. Defects in recombination sharply reduces DNA synthesis at later stages (= DNA arrest). In order to explore the possible function of gene 4 1 in DNA recombination, we sought to isolate a DNA-arrest mutation in gene 4 1 .The pBR322derived plasmid, p415, carries T4 gene 4 1 (Figure 1). Western blotting revealed that p415- encoded gp41 accumulates in the cell, although about 10-fold less than in cells infected with wild-type T4 (data not shown). gp41 was indetectable when cells contain- ing no plasmid were infected with T4 u m N 8 1 (gene 4 1 - ) , but this mutant can grow like wild type on cells carrying p415. Therefore, we used T4 urnN8I to char- acterize plasmid-borne gene 4 1 in the experiments de- scribed below.
After incubation of p415 DNA with hydroxylamine to induce mutations, the DNA was used to transform E . coli MHl cells. Clones recovered in the presence of ampi- cillin were examined for reduced ability to support plaque formation by a m N 8 I phage. Clones with re- duced activity were further examined for the incorpo- ration of ['HI thymidine into the TCA-insoluble fraction of MHl cells infected with umN81 phage. One clone showed arrested DNA synthesis. The plasmid DNA, pHA153, recovered from this clone was again used to transform MHl cells and to reexamine T4 DNA synthe-
sis. The results (see below) confirmed that the abnormal DNA synthesis was attributable to the mutation in the plasmid.
To map the mutation, we successively displaced por- tions of wild-type p415 DNA with the corresponding fragment of pHA153 DNA using restriction enzymes, transformed MH1 cells with the reconstructed plasmids, and examined the transformants' ability to support umN81 phage DNA synthesis. The results are summa- rized in Figure 1. pHAl53 has a 1.2-kb insertion in the gene 4 1 coding region flanked by Sac1 and HindIII(3) (see MATERIALS AND METHODS), but this insertion was
found not to be responsible for the DNA arrest (Figure 1). The chimeric plasmid, p153EH, had the shortest tested fragment from pHA153 causing the arrest of DNA synthesis (Figure 2). The EcoRI-SucI fragment of p153EH DNA was cloned in a phagemid, sequenced by the chain termination method using Tuq DNA polym- erase ( INNIS et al. 1988), and found to have only one base change: the original G at the 17th position up- stream of HindIII(2) in Figure 1 became A (eliminat- ing the PstI site from p415), producing Ala -+ Thr
(Figure 3). Thus, the mutation altered gp41 by a single amino acid change. We named this missense mutation rrh (recombination-deficiency of replicative DNA helicase).
Ultraviolet (W) sensitivity
and
the frequency of genetic recombination: Recovery of phage from UVirradiation and the frequency of genetic recombina- tion relate closely to recombination in the T4 life cycle. To inquire whether rrh impairs recombination, we examined its effects on UV sensitivity and genetic recombination.
DNA Helicase Role in Recombination 249
n r
-
-
p153EH pBR322 s--
c > 8000 U(II 0 4000
.-
2ooo PC
(II
0
0 10 20 30 40
Min at 30"
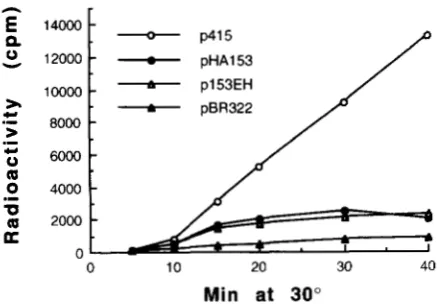
FIGURE 2.-DNA synthesis dependent on plasmid-borne
gene 4 1 . MHl cells harboring various plasmids were grown to
5 X
lo8
cells/ml in M9Amedium supplemented with 20 pg/mlof ampicillin, and were infected at 30" with T4 amN81 phage
at a m.0.i. of 7. T4 DNA synthesis was monitered by incorpo-
ration of [3H] thymidine into the TCA-insoluble fraction in the presence of 5 pg/ml thymidine and 200 pg/ml 2'-
deoxyadenosine (YONESAKI and MINAGAWA 1987). p153EH caused early cessation of DNA synthesis at both 37" and 41"
(data not shown).
p153EH. Phage survival vs. UV dose is 1.5-fold steeper for the rrh allele than for wild-type gene 41.
The frequency of recombination was measured by crossing two different rIIA mutants in an amN81 genetic background (Table 1). In cells harboring p415, the 711' recombinant frequency was 5-6%. In cells harboring p415AHlH3, the frequencywas reduced to 3%. The rrh
allele in pHA153 and in p153EH produced a frequency
Effects on burst size are also shown in Table I. As ex- pected for the DNA-arrest phenotype, rrh results in a low burst size, though significantly higher than that of a null mutation of gene 41. Both pHA153 and p153EH sup- ported plaque formation by amN81 at efficiencies of plating of 0.5-0.8, the plaques being smaller than those on p415. These observations are consistent with the low burst sizes in cells carrying pHAl53 or p153EH.
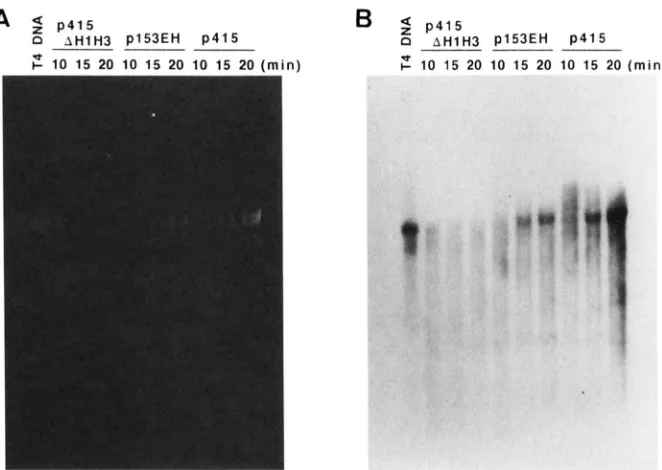
Formation of concatemeric DNA: ssDNA longer than one genomic length of T4 DNA (concatemeric DNA) can be produced by DNA recombination. To obtain evi- dence that the rrh mutation impairs recombination, we analyzed concatemer formation. DNA extracted from amN8l-infected MH1 cells harboring various plasmids was alkalidenatured, electrophoresed through 0.25% agarose gels containing 1 M urea, and stained with
ethidium bromide (Figure 5). The two complementary single strands of T4 DNA migrated at different rates (Fig- ure 5A), demonstrating the ability of this gel system to resolve ssDNAs as long as 171 kb. After the DNA was transferred from the gel onto a nylon membrane, it was detected by southern blotting (Figure 5B). The intensity
of radioactivity was quantitated by densitometry of the X-ray film, and the fraction of concatemeric DNA to of 2-3%.
total DNA for each lane is summarized in Table 2. When the phage-infected cells harbored p415, abundant con- catemeric DNA appeared by 10 min after infection and continued through 20 min, while genome-length T4 DNA appeared at 15 min and became more abundant at 20 min. A control experiment in which cells did not carry a functional gene 41 (p415AHlH3) showed that both concatemeric and genome-length T4 DNAs were poorly accumulated.
On the other hand, when infected cells harbored p153EH, genome-length T4 DNA appeared at 15 min and increased less prominently at 20 min than with p415. Concatemeric DNA was discernible at 10 through 20 min but its fraction among the total DNA synthesized is smaller than that for p415.
DISCUSSION
The rrh mutation in bacteriophage T4 gene 41 causes DNA arrest, diminishes recovery from UV damage, di- minishes genetic recombination, and impairs the for- mation of concatemeric DNA. These phenotypes are qualitatively and quantitatively similar to those for mu- tants defective in recombination-promoting genes such as uvsX (DNA synaptase) and uvsY (enhancer of UvsX recombinase activity) (CUNNINGHAM and BERGER 1977; YONESAKI et al. 1984; YONESAKI and MINAGAWA 1989). Therefore, the results strongly suggest that gene 41
plays a role in T4 recombination and also support our working hypothesis that gene 59 contributes to DNA recombination via the function of gene 41 (YONESAKI 1994).
The T4 primosome consists of DNA helicase plus a DNA primase (gp61). It stimulates the movement of a replication fork and synthesizes an RNA primer essential for the initiation of lagging-strand DNA synthesis (NOSSAL and ALBERTS 1983). I n vitro studies show that gp41 and gp61 are tightly linked in their functions at a replication fork: gp41 enhances RNA primer synthesis by gp61 (
CHA
and ALBERTS 1986) and gp61 stimulates theDNA helicase activity of gp41 (RICHARDSON and NOSSAL
1989). However, null mutations in these genes have dif- ferent phenotypes: a null mutation in gene 41 impairs both origindependent and recombination-dependent initiation, while a null mutation in gene 61 impairs only origindependent initiation, and causes delay of DNA replication. A gene 61 mutation increases recombina- tion fivefold to tenfold (CUNNINGHAM and BERGER 1977). In spite of their cooperation at a replication fork, gp41 and gp61 do not form a stable complex in solution (NOSSAL and ALBERTS 1983). This suggests that gp41 ex-
250 T. Yonesaki
358 360 378 388
- - - L W T A A Q U G K Q A W O S S D U N M S D I A E S A G L P A T - - -
-I
uus 791
T T
FIGURE %-hino acid changes caused by rrh and uus79. A region is shown spanning residues 350-380 of the 475 amino acids in gp41. Both rrh and uus79 cause G --z A changes (at nucleotides 1060 and 1135 in gene 4 1 , respectively) and produce Ala --z
Thr substitutions at amino acids 354 and 379, respectively.
UV dose (sec)
I?
L
0
>
.-
3
tn
0 1 0 2 0 3 0
100
10
1
1
TABLE 1
Recombination frequency and burst size supported by plasmid-borne gene 41
No. of Average Average r+
Plasmid exps. burst size frequency (%)
p415AHlH3 2 7.5 3.0 p415 3 265 5.8
pHA153 2 41 2.6
p153EH 2 32 2.2
MHl cells harboring various plasmids were grown to 5 X 10’
cells/ml in LB broth supplemented with 20 pg,/ml ampicillin and infected at 37” with amN8I rM36 and amN8I r221 phages at an m.0.i. of 5 each. After 1-hr incubation, total progenies were assayed on CR63 ( s u p D ) cells and numbers of rZp recombinants were as- sayed on CR63(hh) cells. Burst size is the ratio of progeny to number of infected cells.
mutants can be explained as follows. In the presence of gp61, only some gp41 participates in recombination, because the rest is sequestered by gp61. Accordingly, a null mutation in gene 61 may allow all gp41 to act in recombination, resulting in more frequent recom- bination and restricting recombination-dependent initiation of only leading-strand synthesis (lagging- strand initiation failing in the absence of a functional gene 6 1 ) .
This hypothesis for the functional states of gp41 can also explain the opposite effects of two different gene 41 missense mutations, uvs 79 and rrh, on recombination and replication. The uvs79 mutant resembles a gene 61- mutant in its increased frequency of recombination (CUPIDO et al. 1980) and slight delay of DNA replication (VAN MINDERHOUT et al. 1978). The uvs 79 mutation may
plasmtd
p415
p415
pHA153
pHA153
p153EH
p153EH
arnN81
+
arnN8 1
+
amN8 1
FIGURE 4 . ” w inactivation. T4 amN81 or wild- type phages
(lo’
pfu/ml in Tris dilution buffer) were irradiated at 1.0 J m-‘ sec” and plated on EHA agar at 37” with MHl host cells harboring various plasmids.impair the interaction of gp41 and gp61, leaving more gp41 free from gp61 like a gene 61- mutation and thus stimulating recombination. In contrast, because rrh al- lows primary DNA replication, the activity of gp41 in the primosome seems unaffected, while its activity in recom- bination is impaired. This mutation is likely to increase the interaction of gp41 with gp61 and to leave less gp41 free. This explanation may be supported by the finding that the rrh and uvs79 sites are very close; each causes a single amino acid change from Ala to Thr at positions (354 and 379, respectively) which are only 25 amino acids apart (Figure 3 ) . If a region containing these amino acids is essential for a gp41 molecule to interact with a gp61 molecule, it seems probable that mutations such as rrh and uvs79 have contrasting effects on the interaction of the two proteins. To clarify this possibility, experiments are required such as the isolation of sup- pressor mutations in gene 61 or biochemical studies of protein-protein interactions.
What is the role of gene 41 in recombination? Genes 46 and 4 7 are essential for DNA recombination and may encode or control an exonuclease responsible for ex- panding an ssDNA nick into a gap (PRASHAD and HOSODA 1972). However, the ssDNA in this gap would not be available for strand transfer by UvsX protein (YONESAKI
DNA Helicase Role in Recombination 25 1
FIGURE 5.-Agarose gel electrophoresis of intracellular T4 DNA under denaturing conditions. MH1 cells (5 X 10' cells/ml in LB supplemented with 20 pg/ml ampicillin) harboring the plasmid indicated in the figure were infected at 37" with T4 amN8I phage at a m.0.i. of 7. At each time, an 0.5ml aliquot was centrifuged for 5 min at 5,000 X gand the infected cells were resuspended in 0.1 ml of 50 mM TrisCI (pH 7.8), 0.1 M NaCI, 10 mM EDTA (pH 8.0) and 0.2% SDS. The suspension was incubated at 37" for 2 hr with proteinase K (500 pg/ml), and the DNA was then denatured with 0.3 M NaOH, loaded onto wells formed in 0.25% agarose in TAE buffer containing 1 M urea, and electrophoresed at 4" for 38 hr at 0.74 V/cm. Urea was added to avoid renaturation of
DNA (OKADA and SHIMURA 1980). (A) The gel was stained with ethidium bromide. (B) The DNA in the same gel was transferred to a nylon membrane with 1.5 M NaCl and 0.5 M NaOH after depurination by washing the gel with 0.25 N HCI. T4 DNA sequences were detected by hybridization with T4 DNA probes. The probe DNA was prepared by sonicating T4 phage DNA, labeling its 5' end with "P, and heatdenaturating the labeled DNA just before use.
TABLE 2
Accumulation of concatemeric DNA
Percent of total DNA in concatemer fraction
Plasmid 10 min 15 min 20 min
p415AHIH3 5.8 7.4 7.8
p153EH 9.9 9.8 8.6
p415 30.4 26.2 20.4
Each lane on the X-ray film shown in Figure 5B was scanned with an image scanner (Epson GT-6000) and the incorporated images were processed by the NIH image program for densitometry of the radioactive intensities in concatemeric DNA and in total DNA.
T4 DNA.
Concatemeric DNA is that migrating slower than genome-length
protein, this 3' end could act as a primer for chain elon- gation by DNA polymerase to initiate recombination- dependent DNA replication (LUDER and MOW 1982).
Although our interpretation seems plausible, it needs more direct evidence, namely that ssDNA generated by gene 41 helicase can be preferentially and/or selectively used for DNA recombination.
We thank J. DRAKE at the National Institute of Environmental Health Sciences for critical reading and invaluable help with the manuscript. We also thank M. SUGITA for her critical reading of this manuscript. This work was supported in part by a Grant-in-Aid to T.Y. from the Ministry of Education, Science, and Culture ofJapan and in part by grant GM24020 to B. ALRERTS at the University of California, San Francisco, during a visit to his laboratory.
LITERATURE CITED
CHA, T.A., and B. M. ALRERTS, 1986 Studies of the DNA helicase- RNA p r i m a e unit from bacteriophage T4. J. Biol. Chem. 261:
CUNNINGHAM, R. P., and H. BERCER, 1977 Mutations affecting recom- bination in bacteriophage T4D. I. Pathway analysis. Virology 8 0
CUPIDO, M.,J. GRIMRERCEN and B. DE GROOT, 1980 A bacteriophage T4 mutant defective in both DNA replication and replication repair. Mutat. Res. 7 0 131-138.
FORMOSA, T., and B. M. ALRERTS, 1986 Purification and characteriza- tion of the T4 bacteriophage uvsX protein. J. Biol. Chem. 261:
FREFSE, E. B., and E. FREFSE, 1964 Two separable effects of hydroxy- lamine on transforming DNA. Proc. Natl. Acad. Sci. USA 5 2
GAUSS, P., K. PARK, T. E. SPENCER and K. J. HACKER, 1994 DNA helicase requirements for DNA replication during bacteriophage T4 in- fection. J. Bacteriol. 1 7 6 1667-1672.
HASHIMOTO, K., and T. YOSES~W, 1991 The characterization of a com- plex of three bacteriophage T4 recombination proteins, uvsX protein, uvsY protein, and gene 32 protein, on single-stranded DNA. J. Biol. Chem. 266: 488.3-4888.
INNIS, M. A., K. B. MYAMRO, D. H. GEL.FAVD and M. A. BROW, 1988 DNA sequencing with Thtrmus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc. Natl. Acad. Sci. USA 85: 9436-9440.
LUDER, A,, and G . Moslc, 1982 Two alternative mechanisms for ini- tiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc. Natl. Acad. Sci.
MOSIG, G., 1987 The essential role of recombination in phage T4 growth. Annu. Rev. Genet. 21: 347-371.
nos.,\^., N. G., and R. M. AI.RERTS, 1983 Mechanism of DNA repli- cation catalyzed by purified T4 replication proteins, pp.71-81 7001-7100.
67-82.
6107-6118.
1289-1297.
252 T. Yonesaki
in Bacteriophage T4, edited by C. K. MATHEWS, E. M. KUTTER,
G. MOSIG and P. B. BERGET. American Society for Microbiology, Washington, D.C.
OKADA, IC, and Y. SHIMURA, 1980 Arrangement of the single-stranded fragments in E. coli bacteriophage BF23 DNA. Gene 8: 345-368.
PRASHAD, N., and J. HOSODA, 1972 Role of genes 46 and 47 in bac- teriophage T4 reproduction. 11. Formation of gaps on parental
DNA of polynucleotide ligase defective mutants. J. Mol. Biol. 70:
RICHARDSON, R. W., and N. G. NOSSAL, 1989 Characterization of the bacteriophage T4 gene 41 DNA helicase. J. Biol. Chem. 264:
VAN MINDENOUT, L., J. BRIMBERGEN and B. DE GROOT, 1978 Non- essential UV-sensitive bacteriophage T4 mutants affecting early
DNA synthesis: a third pathway of DNA repair. Mutat. Res. 52:
313-322. 617-635.
4725-4731.
YONESAKI, T., 1994 The purification and characterization of gene
59 protein from bacteriophage T4. J. Biol. Chem. 269: 1284- 1288.
YONESAKI, T., and T. MINACAWA, 1985 T4 phage gene uusX product catalyzes homologous DNA pairing. EMBO J. 4 3321-3327.
YONESAKI, T., and T. MINAGAWA, 1987 Studies on the recombination genes of bacteriophage T4: suppression of U U S X and UUSY mu- tations by uvsWmutations. Genetics 115: 219-227.
YONESAKI, T., and T. MINAGAWA, 1989 Synergistic action of three re- combination gene products of bacteriophage T4, uvsX, UVSY, and gene 32 proteins. J. Biol. Chem. 264: 7814-7820.
YONESAKI, T., J. M~VAZAK~ and T. MINAGAWA, 1984 Genetic studies on non-lethal recombination gene mutants of bacteriophage T4. Mem. Fac. Sci. Kyoto Univ. Ser. Biol. 9: 87-96.