Effects of Antibiotics on
Bacterial Growth and
Protein Synthesis:
Instructor’s Manual
I. Purpose and Concepts Covered...1
II. Inhibition of Bacterial Growth...2
A. Preparation for the Laboratory...2
B. Protocol ...3
C. Expected Results...4
D. Troubleshooting ...5
III. Inhibition of Translation...6
A. Preparation for the Laboratory...6
B. Supplemental Protocol Information ...8
C. in vitro Transcription/Translation Protocol ...9
D. Expected Results...11
E. Troubleshooting ...12
F. Additional Resources...13
IV. Alternative Protocol for Laboratories Without a Luminometer...14
A. Expected Results of Alternative Protocol ...14
B. Additional Discussion Questions for Alternative Protocol ...15
V. Supplier and Ordering Information...15
I. Purpose and Concepts Covered
The purpose of this experiment is to demonstrate the differential effects of various antibiotics on bacterial growth and translation in an in vitro prokaryotic protein expression system (S30 E. coli extract).
This laboratory exercise provides an opportunity to cover the following topics: • translation/protein synthesis
• drug screening to identify new bacteriostatic agents (to apply information learned here to a new concept)
• the relationship between in vitro and in vivo results in experimental design
This instructor’s manual is available online only.
This teaching resource is made available free of charge by Promega Corporation. Reproduction permitted for noncommer-cial educational purposes only. Copyright 2007 Promega Corporation. All rights reserved.
II. Inhibition of Bacterial Growth Materials Required
Mueller Hinton broth* (dehydrated medium for reconsitution with water into agar or broth; BBL Microbiology Cat.# 211443*
Mueller Hinton agar* (BBL Microbiology Cat.# 211438) Distilled water
Petri dishes (Fisher Scientific Cat.# 08-757-9B)
E. coli cultures with an optical density at 600nm in the range of 0.08 to 0.1 for each student or group of students
Antibiotic disks (see supplier and ordering information at back of manual) Small beaker with alcohol and forceps
Sterile cottom swabs Bunsen burner Metric ruler
Markers to label plates
*Mueller Hinton medium is standard for this type of test, but LB medium can be substituted.
II.A. Preparation for the Laboratory
1. Prepare Mueller Hinton broth to grow overnight culture of E. coli. Compositions are given below.
2. Prepare Mueller Hinton agar plates. Prepare two plates per student or group of students.
3. Prepare cultures ofE. coli. Incubate with shaking at 37°C until the culture has an optical density at 600 nm of 0.08–0.1 or until the turbidity matches that of a McFarland 0.5 standard, which is used as a turbidity standard to prepare bacterial cultures. On the day of the lab, dispense the cultures into sterile tubes so that there is at least 5 ml of culture per student or group of students.
4. Ensure that each student or group of students has a Bunsen burner, small beaker with alcohol and forceps, sterile cotten swabs, metric rulers and markers to label plates.
5. Prior to the laboratory exercise, equilibrate the Mueller Hinton plates to room temperature.
Preparation of Agars and Broths
Mueller Hinton broth
Use premixed Mueller Hinton medium, and prepare as instructed by the manufacturer. Autoclave at 121 °C and 20 psi for 30 minutes. If you do not have access to an auto-clave, boil the medium or cook it in a pressure cooker for 1 hour.
Mueller Hinton agar plates
Prepare Mueller Hinton broth as described above but add 15 g of agar per liter. Autoclave, and allow the medium to cool to approximately 55 °C. Pour into plates. Store the plates at 4 °C.
LB broth
Prepare LB broth by combining 5 g yeast extract, 10 g Bacto-tryptone and 5 g NaCl. Add water to bring the final volume to 1 liter. Mix and autoclave at 121 °C and 20 psi for 30 minutes.
LB agar plates
Prepare LB broth as described above but add 15 g of agar. Autoclave and allow the medium to cool to approximately 55 °C. Pour into plates. Store the plates at 4 °C.
II.B. Protocol
Do not handle antibiotic disks if you are allergic to the antibiotic.
Day 1
1. Inoculate one Mueller Hinton agar plate with E. coli. Dip a sterile cotton swab into the overnight culture of E. coli, and wipe off any excess on the inside of the tube. Inoculate a Mueller Hinton agar plate by streaking the entire surface of the plate with the swab, turning the plate 90°, swabbing a second time, turning the plate 45° and swabbing a third time. Run the swab around the circumference of the plate. Be sure to cover the entire plate. Discard the swab.
2. Repeat Step 1 using a fresh sterile swab to inoculate the second Mueller Hinton agar plate.
3. Allow the plates to dry for 5 minutes before placing the antibiotic disks on the plate surface.
4. Apply four antibiotic disks to the agar surface of one plate using sterile for-ceps. To sterilize the forceps, dip them in alcohol, letting the excess drip into the beaker, and pass them through the Bunsen burner flame. Allow the alco-hol to burn off. Be sure to space the disks at least 4–5 cm apart to prevent overlapping zones of growth inhibition. Press each disk gently with sterile for-ceps so that the disk makes good contact with the surface of the agar.
Note:Each antibiotic disk will be marked with an abbreviation (Table 1). The blank disk has no markings.
5. Repeat Step 4 to apply the other two antibiotic disks and the blank disk to the second Mueller Hinton agar plate.
6. Cover the plates. Label the plates with your name, date and organism name. 7. Invert the plates, and incubate them at 35–37 °C for 18–24 hours.
Day 2
1. Remove the plates from the incubator. With a metric ruler, measure the zone of inhibition, the distance between the edge of the disk and edge of bacterial growth, at the widest point.
Note:Results may be easier to read if plates are placed on a black back-ground or a light box.
2. Record the results in Table 1. Record whether the compound inhibited bacterial growth.
II.C. Expected Results
Zones will vary based on growth phase of bacteria when plated, depth of agar and environment. However students should see zones around all of the antibiotics tested in this laboratory. Table 2 provides guidelines for describing E. coli bacteria as susceptible, intermediate or resistant.
Discussion
1. Which compounds were the most effective in inhibiting growth of E. coli based on the width of the zone of inhibition? Which were the least effective? 2. Based on the mechanisms of action listed in Table 3, why was growth
inhibited? Can you explain why some of the antibiotics tested were less effective than others at inhibiting growth of E. coli ? Would you have expected similar results if you had tested S. aureus in your assay? Table 3. Modes of Action.
Compound Mechanism of Action
Ampicillin
Inhibits cell wall synthesis by inhibiting formation of the peptidoglycan cross-link.
Chloramphenicol Inhibits prokaryotic peptidyl transferase. Streptomycin
Inhibits prokaryotic peptide chain initiation, and induces mRNA misreading.
Tetracycline
Inhibits prokaryotic aminoacyl-tRNA binding to the ribosome small subunit.
Erythromycin Inhibits prokaryotic peptide chain initiation.
Neomycin Inhibits prokaryotic translocation through the ribosome large subunit. Cycloheximide Inhibits eukaryotic peptidyl transferase.
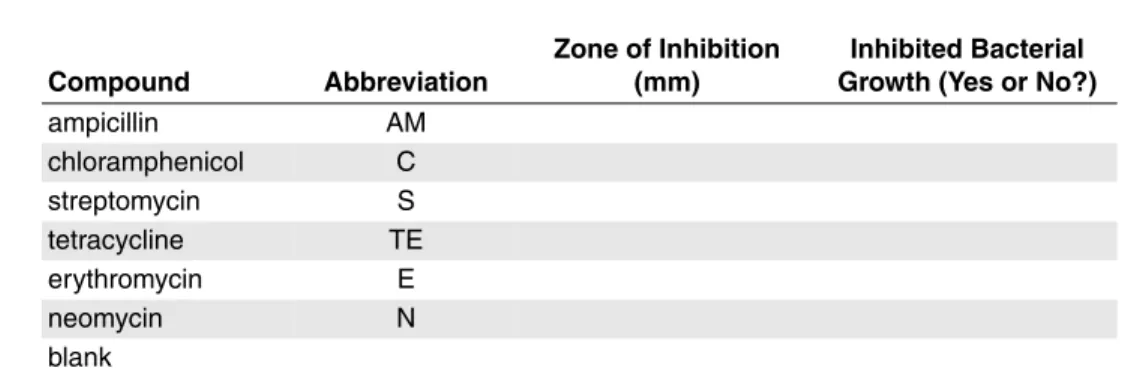
Note:This is Table 2 in the Students’ Laboratory Manual. Table 1. Effect of Compounds on Bacterial Growth.
Compound Abbreviation
Zone of Inhibition (mm)
Inhibited Bacterial Growth (Yes or No?)
ampicillin AM chloramphenicol C streptomycin S tetracycline TE erythromycin E neomycin N blank
Table 2. Diameter of Zones of Inhibition forE. coli.
Compound Susceptible Intermediate Resistant
ampicillin (10 µg) 14 mm or more 12–13 mm 11 mm or less
chloramphenicol (39 µg) 18 mm or more 13–17 mm 12 mm or less streptomycin (10 µg) 15 mm or more 12–14 mm 11 mm or less tetracycline (30 µg) 19 mm or more 11–15 mm 14 mm or less erythromycin (15 µg) 18 mm or more 14–17 mm 13 mm or less
neomycin (30 µg) 17 mm or more 13–16 mm 12 mm or less
II.D. Troubleshooting
Symptoms Discussion
No growth on bacterial plates Plates were too warm or cold when bacteria
were streaked. Be sure to equilibrate plates to room temperature before use.
Plates were not properly prepared or were old. Use freshly prepared plates that were made with unexpired components. Incubator was not at 37 °C. Double-check the incubator temperature.
Students streaked plates with a swab that had been dipped in alcohol. Be sure that students understand the procedure.
Colonies appearing within the Plate is contaminated with other bacteria
lawn ofE. coli on the plates or a fungus. Be sure students follow sterile techique when plating E. coli and antibiotic disks.
Surface of plate is covered with Plates were too warm or cold when plated,
water and condensation covered the agar surface.
Equilibrate plates to room temperature, and incubate upside down in the incubator. Condensation on top of plate accumulated on agar surface during incubation. Incubate plates upside down in the incubator. No zones of inhibition detected Antibiotic disks were too old. Check
around any disks expiration dates. Use fresh disks.
Zone of inhibition detected around Students dipped disk in alcohol. Make sure
blank control disk that students understand the procedure.
Zones of inhibition are widely Make sure that plates are poured to a
varying uniform depth. In the clinical setting the
standard depth is 4 mm. The depth of the agar can affect the concentration of the antibiotic as it diffuses from the disk.
III. Inhibition of Transcription/Translation Materials Required for the Laboratory Exercise
E. coli S30 Extract System for Circular DNA (Promega Cat.# L1020)
Steady-Glo®Luciferase Assay System (Promega Cat.# E2510)
pBESTluc™ DNA (1µg/µl)
chloramphenicol (Sigma Aldrich Cat.# C7795
streptomycin sulfate salt (Sigma Aldrich Cat.# S0890) tetracycline hydrochloride (Sigma Aldrich Cat.# T7660) erythromycin (Sigma Aldrich Cat.# E5389)
neomycin solution (Sigma Aldrich Cat.# N1142)
cycloheximide ready made solution (Sigma Aldrich Cat.# C4859) nuclease-free water
pipettes and pipette tips 1.5 ml microcentrifuge tubes distilled water at room temperature heat block or water bath at 37 °C
Each E. coli S30 Extract System contains sufficient reagents for 30 in vitro translation reactions. This exercise requires enough reagents for 10 reactions per student or group of students. The E. coli S30 Extract System contains enough pBESTluc™ DNA for 20 reactions.Note:Be sure to request an additional vial of the pBESTluc™ DNA from Promega to ensure that there is enough DNA template for your reactions.
Each Steady-Glo®Luciferase Assay System (Cat.# E2510, 10 ml) includes enough reagents for 100 luciferase assays. Each student or group of students will be performing 10 luciferase assays. This system is also available in a 100 ml size (Cat.# E2520), which includes enough reagents for 1,000 assays.
We recommend using the Steady-Glo®Luciferase Assay System to measure light output. The Luciferase Assay Reagent supplied with the S30 Extract System has a half-life of 10 minutes, whereas the Steady-Glo®Reagent has a half-life of approximately 5 hours, resulting in less than 13% loss of luminescence per hour. The longer half-life of the Steady-Glo®Reagent allows students to add the reagent at the bench before proceeding to the luminometer. If the supplied Luciferase Assay Reagent were used, students would be required to add the reagent to each tube immediately before measuring luminescence.
III.A. Preparation for the Laboratory
Prepare stock solutions for each compound as directed in Table 4. Prepare the working solution by diluting the stock solution for each compound to the indicated concentration in nuclease-free water. Store at the appropriate temperature.
Add 10 µl of nuclease-free water to 10 µl of pBESTluc™ Vector to dilute the vector 1:2 for a final concentration of 0.5 µg/µl. Mix, and dispense the vector into the appropriate number of aliquots for the number of students or groups of students. Store the dispensed vector on ice. Store any unused vector at –20 °C.
Up to 4 hours before the lab begins, prepare the Steady-Glo®Reagent by transfer-ring the contents of one bottle of Steady-Glo®Buffer to one bottle of Steady-Glo® Substrate. Mix by inversion until the substrate is thoroughly dissolved. Store the reagent at room temperature until use.
Note:Since luciferase activity is temperature-dependent, the temperature of the Steady-Glo®Reagent should be held constant while quantitating luminescence. This is achieved most easily by using Steady-Glo®Reagent equilibrated to room temperature, which is near the temperature optimum of luciferase. To equilibrate the Steady-Glo®Reagent to room temperature, place the tube containing the Steady-Glo® Reagent into a container of room-temperature water for 30 minutes prior to use. If cold reagent is used to detect luciferase activity, luminescence will slowly increase during the experiments as the reagent warms, introducing inaccuracies. High temperatures cause an increase in luminescence, but the signal becomes less stable. This can occur if the reagent is too warm or if the luminometer produces excess heat within the reading chamber.
Storage Conditions
E. coli S30 Extract System for Circular DNA:Store all components at –70 °C. The product is sensitive to CO2(avoid prolonged exposure to CO2sources such as dry ice) and multiple freeze-thaw cycles, which may have an adverse affect on activity or performance. Before use, thaw the components on ice. The S30 extract is temperature-sensitive, so thaw the S30 extract just prior to use.
Steady-Glo®Luciferase Assay System:Store the lyophilized Steady-Glo® Substrate at –20 °C. The substrate may also be stored at 4 °C for up to one month. Store the Steady-Glo®Buffer below 25 °C. Storage at room temperature is
recommended to eliminate the need for temperature equilibration when the reagent is reconstituted. Use the reconstituted Steady-Glo®Reagent on the same day it is prepared, or store at –20 °C for up to 2 weeks. Storing the prepared reagent at room temperature will result in a 7% loss of luminescence per 8 hours, 10% loss per 24 hours at 4 °C and 8% loss per 2 weeks at –20 °C. The reagent may be subjected to up to five freeze-thaw cycles with no effect on potency. Frozen reconstituted reagent should be thawed below 25 °C to ensure reagent performance. Mix well after thawing. The most convenient and effective method for thawing or equilibrating Table 4. Preparation and Storage Conditions for Antibiotics.
Sigma
Cat.# Compound Name Stock Solution Working Solution
Recommended Storage Temperature C7795 Chloramphenicol 20 mg/ml in ethanol 10 mg/ml in water 2–8 °C; use stock solution within
30 days S0890 Streptomycin
sulfate salt
10 mg/ml in 0.9% NaCl 10 mg/ml in water 2–8 °C for up to 1 month, or –20 °C for extended periods. T7660 Tetracycline hydrochloride 10 mg/ml in water 10 mg/ml in water –20 °C
E5389 Erythromycin 20 mg/ml in ethanol 10 mg/ml in water 2–8 °C
N1142 Neomycin solution 10 mg/ml in 0.9% NaCl 10 mg/ml in water 2–8 °C C4859 Cycloheximide
ready made solution
Test Compounds:Recommended storage conditions for the compounds used in these experiments can be found in Table 4 and in the literature supplied by Sigma Aldrich.
Precautions
The lyophilized Steady-Glo®Substrate contains dithiothreitol (DTT) and is therefore classified as hazardous. The reconstituted reagent is not known to present any hazards, as the concentration of DTT is less than 1%. However, we recommend the use of gloves, lab coats and eye protection when working with these or any chemical reagents. Additional information about potential hazards can be found on the
Material Safety Data Sheet supplied with the reagents.
III.B. Supplemental Protocol Information
This protocol requires pipetting volumes as small as 1.0 µl. Failure to pipet accurately can result in a shortage of S30 Extract System components or other reagents. The S30 Extract is the limiting reagent in the S30 E. coli Extract System. The extract can be viscous, so take care to pipet near the top of the liquid level and do not submerge the pipet tip to the bottom of the tube, because small droplets of liquid can adhere to the pipet tip. Consider your students’ levels of experience, and if necessary, provide the students with a short lesson or reminder on how to pipet accurately.
The reaction may be incubated within a temperature range of 24–37 °C. The fastest linear rate of protein synthesis occurs at 37 °C for approximately 1 hour, although the reaction will continue for several hours at a slower rate. Lower temperatures result in a slower rate of synthesis but often extend the time of the linear rate to several hours. Protein yields from the S30 Extract System for Circular DNA vary with the template and conditions used. Typical yields range from 50–250 ng per reaction.
When measuring firefly luciferase activity, background luminescence must be subtracted from all readings. No background is produced by the Steady-Glo®Reagent or S30 extract lacking the pBESTluc™ DNA, so background luminescence is a characteristic of luminometer performance. Some instruments also require verification of linear response at high light levels (consult the instrument manual). In the laboratory protocol, students are instructed to measure background by adding 100 µl of
Steady-Glo®Reagent to 100 µl of distilled water. Substract this background reading from the experimental readings (raw luminescence) to calculate net luminescence.
III.C. in vitro Transcription/Translation Protocol
Use a fresh pipet tip for each reagent addition.
1. Prepare enough master mix for 9 in vitro transcription/translation reactions by combining the following reaction components.
2. Vortex gently, then centrifuge in a microcentrifuge for 5 seconds to bring the reaction mixture to the bottom of the tube.
3. Label eight 1.5 ml microcentrifuge tubes as reactions 1 to 8. Pipet 49 µl of the master mix prepared in Step 1 to each of the tubes.
4. Pipet 1.0 µl of the compound to a tube containing the master mix as follows:
5. Add 1.0 µl of nuclease-free water to tube 8.
Note:Tube 8 does not contain a compound and will act as the positive control. This reaction represents the maximum level of protein synthesis.
6. Vortex each reaction gently to mix. Incubate the reaction at 37 °C for 60 minutes. 7. Stop the reaction by placing the tubes on ice for 5 minutes.
Compound Tube Number
ampicillin 1 chloramphenicol 2 streptomycin 3 tetracycline 4 erythromycin 5 neomycin 6 cycloheximide 7 Component Volume Per Reaction × Number of Reactions1 = Final Volume pBESTluc™ DNA (0.5 µg/µl) 1.0 µl
Amino Acid Mixture Minus Cysteine,
1mM (mix gently prior to use) 2.5 µl Amino Acid Mixture Minus Methionine,
1mM (mix gently prior to use) 2.5 µl S30 Premix Without Amino Acids
(mix gently prior to use) 20 µl
S30 Extract, Circular2
(mix gently prior to use) 15 µl
Nuclease-free water 8.0 µl
Final volume 50 µl
1You will assemble 8 reactions in this exercise but will prepare enough master mix for
9 reactions. This should ensure that you have enough master mix.
2The extract can be viscous, so take care to pipet near the top of the liquid level and do not
submerge the pipet tip to the bottom of the tube, because small droplets of liquid can adhere to the pipet tip.
Detection of Luciferase Protein
1. Label nine microcentrifuge tubes 1 to 9. Add 95 µl of distilled water to tubes 1–8. 2. Add 100 µl of distilled water to tube 9.
3. Add 5.0 µl of each translation reaction to tubes 1–8.
4. Add 100 µl of Steady-Glo™ Reagent that has been equilibrated to room temperature to each tube, and mix by pipetting.
Note:Light intensity is a measure of the amount of luciferase present but also the enzymatic rate, which depends upon temperature. Be sure that the Steady-Glo® Reagent has been equilibrated to room temperature (20–25 °C) for reproducible luciferase assay readings.
5. Place the reaction in a luminometer or multimode instrument capable of measuring luminescence. Measure the luminescence and record the results in Table 5. Consult the appropriate operator's manual for operation of the luminometer, if necessary.
Note:Luminescence is measured in terms of relative light units (RLU).
5. Calculate the net luminescence by subtracting the background luminescence measured in tube 9 from the experimental luminescence in tubes 1–8. Record net luminescence values in Table 5.
6. Calculate the percent of inhibition for each of the compounds using the equation given below. Reaction 8 did not contain an inhibitor, and the light output from this reaction represents the maximum level of protein synthesis.
Net Luminescence in the Presence of Compound
× 100% Net Luminescence in the Absence of Compound
Table 5. Effect of Compounds on in vitro Transcription/Translation. Tube Number Compound Raw Luminescence (RLU) Net Luminescence (RLU) Percent Inhibition of Protein Synthesis 1 ampicillin 2 chloramphenicol 3 streptomycin 4 tetracycline 5 erythromycin 6 neomycin 7 cycloheximide 8 no compound – 9 no compound; no translation reaction –
III.D. Expected Results
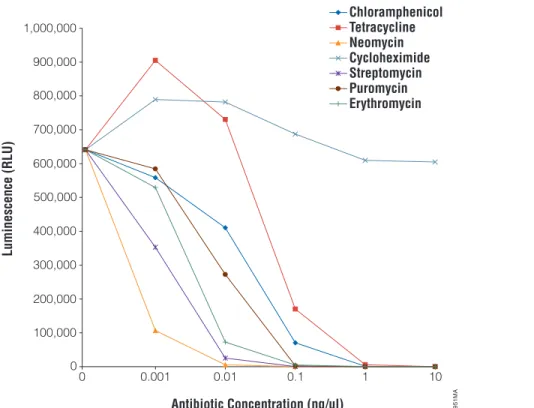
Cycloheximide will not inhibit translation in the E. coli S30 system but does inhibit translation in eukaryotic systems. Puromycin will efficiently inhibit both eukaryotic and prokaryotic translation. The other antibiotics tested (chloramphenicol, streptomycin, tetracycline, erythromycin, and neomycin) only inhibit prokaryotic translation. In experiments performed at Promega, these compounds showed IC50values in the range of 0.1–1.0 ng/µl for the S30 E. coli Extract System. The most potent translational inhibitors are neomycin, followed closely by streptomycin and erythromycin (Figure 1).
Note:A relative light unit is not an absolute unit of measurement. The number of relative light units (RLU) produced in a reaction depends on the exact experimental conditions and can vary significantly, so you should not expect to obtain exactly the same results as shown in Figure 1. In addition, different luminometer manufacturers will define a relative light unit differently. Therefore, different luminometers will often yield different results; the number of relative light units determined with one lumino-meter may be dramatically higher than that with a different luminolumino-meter. This difference affects all measurements taken with a particular luminometer so that, although the light output might be dramatically higher, the background will also be proportionally higher. The net result is that the signal-to-background ratio is consistent.
Figure 1. Representative results.All compounds were diluted to 10 mg/ml, then 1:10 serial dilutions were performed from 1:10 to 1:10,000 (1 mg/ml to 0.001 mg/ml, respectively). One microliter of each serial dilution was added to a 50 µl in vitro translation reaction containing 0.5 µg of pBESTluc™ DNA. Reactions were incubated at 37 °C for 1 hour. Luminescence was measured using the Steady-Glo®
Reagent and a Berthold EG&G MicroLumat Plus plate-reading luminometer (RLU factor = 10.0; integration time = 2.0 seconds).
6951MA 0 100,000 200,000 300,000 400,000 500,000 600,000 700,000 800,000 900,000 1,000,000 10 1 0.1 0.01 0.001 0 Antibiotic Concentration (ng/µl) Luminescence (RLU) Chloramphenicol Tetracycline Neomycin Cycloheximide Streptomycin Puromycin Erythromycin
Supplemental Discussion Questions
1. As bacteria acquire resistance to existing antibiotics, new antibiotics need to be developed to combat antibiotic-resistant bacterial strains. Based on this protocol, design an experiment to screen for new compounds that preferen-tially inhibit bacterial translation and could have antibiotic properties. 2. A population of bacteria exposed to an antibiotic stops growing when the
antibiotic concentration is equal to or higher than the minimum inhibitory concentration (MIC). How could you design an experiment to determine the MIC for the compounds tested today?
3. Some antibiotics are bacteriostatic and inhibit bacterial growth, whereas other antibiotics are bactericidal and cause bacterial cell death. Based on the mode of action of the antibiotics used in this lab (Table 2), which antibiotics would you expect to be bacteriostatic and which bacteriocidal?
4. What results would you expect if you performed the inhibition of transcription/ translation experiments with a rabbit reticulocyte lysate-base transcription/ translation system rather than the bacterial S30 Extract System? Which antibiotics would result in cell death, and which antibiotics would have no effect?
5. E. coli transformed with the pBESTluc™ Vector will express firefly luciferase protein. What DNA features are necessary for expression of this protein from the pBESTluc™ Vector? What RNA features are required?
III.E. Troubleshooting
Symptoms Causes and Comments
No light output from the A component of the in vitro transcription/
in vitro transcription/ translation reaction was omitted. Be sure
translation reaction that reactions were assembled correctly.
S30 Extract lost activity. The S30 Extract is temperature-sensitive. Thaw the extract just before use, and keep on ice while working with the S30 Extract. Minimize the amount of time that the extract is spent on ice. Do not subject the S30 Extract to multiple freeze-thaw cycles. After the initial thaw, quickly refreeze any unused S30 Extract in single-use aliquots in a dry ice-ethanol bath, and store at –70° C.
Low light output from Impurities in the water used to assemble the
the in vitro transcription/ reaction inhibited transcription or translation or
translation reaction degraded the DNA or RNA molecules in the
reaction. Water purity is extremely important. If translation efficiencies are low, examine the quality of the water. Use only nuclease-free water.
S30 Extract lost activity. The S30 Extract is temperature-sensitive. Thaw the extract just before use, and keep on ice while working with the S30 Extract. Minimize the amount of time that the extract is spent on ice. Do not subject the S30 Extract to multiple freeze-thaw cycles. After the initial thaw, quickly refreeze any unused S30 Extract in single-use aliquots in a dry ice-ethanol bath, and store at –70° C.
III.E. Troubleshooting (continued)
Symptoms Causes and Comments
Low light output from The S30 Extract is sensitive to multiple
freeze-the in vitro transcription/ thaw cycles. Do not subject the S30 Extract to
translation reaction multiple freeze-thaw cycles. After the initial
(continued) thaw, quickly refreeze any unused S30 Extract
in single-use aliquots in a dry ice-ethanol bath, and store at –70°C.
Reaction components were not mixed before use. Be sure that students mix each
component well by pipetting before use. S30 Extract lost activity. S30 extract is sensitive to CO2. Do not expose the S30 Extract to CO2sources such as dry ice for prolonged periods of time.
Firefly luciferase protein produced in the transcription/translation reaction lost activity. Luciferase has an optimal reaction temperature near room temperature. Temperatures above 30 °C can cause thermal inactivation. Be sure to equilibrate the Steady-Glo®reagent to room temperature to avoid thermal inactivation of the luciferase protein.
Reactions were not assembled correctly. Be sure that students pipet reaction components accurately. The S30 Extract can be viscous, so be sure that students pipet near the top of the liquid level and do not submerge the pipet tip to the bottom of the tube, as small droplets of liquid can adhere to the pipette tip.
III.F. Additional Resources
Additional information about the Steady-Glo®Luciferase Assay System and E. coli S30 Extract System for Circular DNA can be found at the Promega Web site:
Steady-Glo®Luciferase Assay System:www.promega.com/tbs/tm051/tm051.html
E. coli S30 Extract System for Circular DNA:
IV. Alternative Protocol for Laboratories Without a Luminometer
The alternative protocol is suitable for laboratories without a luminometer or multimode instrument capable of measuring luminescence or for instructors who wish to introduce students to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protocol uses the Transcend™ Non-Radioactive Translation Detection Systems (Cat.# L5070 and L5080) to detect the luciferase protein. Using these systems, biotinylated lysine residues are incorporated into nascent proteins during translation. This biotinylated lysine is added to the translation reaction as a precharged ε-labeled biotinylated lysine-tRNA complex (Transcend™ lysine-tRNA) rather than a free amino acid. After SDS-PAGE and electroblotting, the biotinylated proteins are visualized by binding streptavidin-alkaline phosphatase, followed by colorimetric detection. See the Transcend™ Non-Radioactive Translation Detection Systems Technical Bulletin TB182 for more information. To use the alternative protocol, substitute 1–2 µl of Transcend™ tRNA for the same volume of nuclease-free water. Assemble the reactions as directed in the Student Laboratory Manual, but omit 1–2 µl of nuclease-free water. Add all components except the Transcend™ tRNA, and gently mix the reaction by pipetting while stirring the reaction with the pipette tip. If necessary, spin briefly in a microcentrifuge to return the sample to the bottom of the tube. Add the Transcend™ tRNA after mixing, and incubate the reactions as directed.
IV.A. Expected Results of Alternative Protocol
When using this altenative protocol, the synthesized firefly luciferase protein will be separated by size using SDS-PAGE. The luciferase migrates at 61 kDa. An apparent internal translation start results in a second major gene product of 48 kDa. This plasmid also contains the gene for ampicillin resistance (β-lactamase).β-lactamase may appear as a faint band migrating at 31.5 kDa. See Figure 2 for typical results. Many eukaryotic genes contain sequences within the coding region that can function as ribosomal binding sites when they precede a methionine codon. The presence of such internal sequences can result in internal translation initiation and the synthesis of potentially undesired truncated proteins in the prokaryotic system. An example of this can be seen when expressing the firefly luciferase gene in the E. coli S30 Extract System. The firefly luciferase gene contains 14 methionine codons, several of which are preceded by potential ribosome binding site (RBS) sequences and produce truncated translation products.
0692T A07_4A luciferase (61kDa) luciferase internal start (48kDa) β-lactamase (31.5kDa) 1 2 3 kDa 97.4 – 69.0 – 46.0 – 30.0 –
Figure 2. Coupled in vitro transcription/translation of circular DNA templates using the E. coli S30 Extract System for Circular DNA.Five microliters of each 50 µl reaction mix was loaded in each lane. Lanes 2 and 3 show protein products synthesized from pBESTluc™ DNA. Full-length luciferase migrates at 61 kDa. An apparent internal translation start results in a second major gene product of 48 kDa.β-lactamase migrates at 31.5 kDa. Lane 1 shows the molecular weight markers; lanes 2 and 3 are duplicate translation reactions.
IV.B. Additional Discussion Questions for Alternative Protocol
1. Full-length firefly luciferase protein has a molecular weight of 61 kDa. The firefly luciferase gene contains 14 methionine codons, several of which are preceded by potential ribosome binding site (RBS) sequences. Apply this knowledge to explain the 48 kDa translation product that can be observed after SDS-PAGE. 2. How are proteins separated by size in an SDS-polyacrylamide gel?
V. Supplier and Ordering Information Ordering Information
Product Cat.#
E. coli S30 Extract System for Circular DNA L1020
Provides enough control plasmid for 20 student reactions. (Each student group will perform 10 reactions.) Be sure to request an adiditional vial of pBESTluc™ DNA to ensuret hat there is enough DNA template for your reactions.
Steady-Glo®Luciferase Assay System E2510
Provides enough reagent for 100 luciferase assays. (Each student group will be performing 10 luciferase assays.)
Transcend™ Non-Radioactive Translation Detection System L5070
For Laboratory Use. For laboratories that do not have access to a luminometer or multimode instrument that can mea-sure luminescence. Also, instructors who wish to introduce students to SDS-PAGE can use this system.
Nuclease-Free Water P1193
For Laboratory Use.
Promega Training Support Program: Discounts for Educators in the United States
To order Promega products for your teaching laboratory (U.S. only) at a significant
educational discount, visit:www.promega.com/us/trainingsupport/default.htm
Materials Required from Vendors Other than Promega
Product Vendor Cat.#
Mueller Hinton Agar BBL Microbiology 211438
Mueller Hinton Broth (dehydrated powder) BBL Microbiology 211443
Ampicillin antibiotic disks (10 µg) BBL Microbiology 230705
Chloramphenicol antibiotic disks (30 µg) BBL Microbiology 230733
Streptomycin antibiotic disks (10 µg) BBL Microbiology 230942
Tetracycline antibiotic disks (30 µg) BBL Microbiology 231344
Erythromycin antibiotic disks (15 µg) BBL Microbiology 230793
Neomycin antibiotic disks (30 µg) BBL Microbiology 230882
Blank paper disks BBL Microbiology 231039
Ampicillin sodium salt Sigma-Aldrich A2084
Chloramphenicol Sigma-Aldrich C7795
Streptomycin sulfate salt Sigma-Aldrich S8090
Tetracycline hydrochloride Sigma-Aldrich T7660
Erythromycin Sigma-Aldrich E5389
Neomycin solution Sigma-Aldrich N1142
Cycloheximide ready made solution Sigma-Aldrich C4859
Petri dishes (10 cm) Fisher Scientific 08-757-9B
Products may be covered by pending or issued patents or may have certain limitations. Please visit our Web site for more information.