organic papers
Acta Cryst.(2005). E61, o1769–o1770 doi:10.1107/S1600536805014844 Yang Liet al. C
14H10O5
o1769
Acta Crystallographica Section E
Structure Reports
Online
ISSN 1600-5368
2-(2,4-Dihydroxybenzoyl)benzoic acid
Yang Li,a* Feng-Yan Ge,a Li-Gong Chen,aChuan-Ming Dong,aXi-Long Yan,aEr-Hong Duan,bTao Zeng,aYue-Cheng Zhangaand Guo-Yi Baic
aCollege of Pharmaceuticals and Biotechnology,
Tianjin University, Tianjin 300072, People’s Repulic of China,bResearch and Development Center for Petrochemical Technology, Tianjin University, Tianjin 300072, People’s Repulic of China, andcCollege of Chemistry and Environmental Science, Hebei University, Baoding 071002, People’s Repulic of China
Correspondence e-mail: liyang777@tju.edu.cn
Key indicators
Single-crystal X-ray study T= 294 K
Mean(C–C) = 0.003 A˚ Rfactor = 0.049 wRfactor = 0.146
Data-to-parameter ratio = 14.3
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
#2005 International Union of Crystallography Printed in Great Britain – all rights reserved
In the title structure, C14H10O5, the angle between the planes formed by the 2,4-dihydroxybenzoyl and o-benzoic acid moieties is 87.12 (4). In addition to an intramolecular O—
H O hydrogen bond, intermolecular O—H O hydrogen bonds (H O = 1.76 and 1.89 A˚ ) connect molecules to form a two-dimensional network parallel to (101).
Comment
The title compound, (I), is an intermediate in the synthesis of fluorescein and was first prepared by Baeyer (1876) and also by Bollmann (1922) in his study of resorcinbenzein. Further details about the title compound and its derivatives have been reported (Orndorff & Adamson, 1918; Orndorff & Kelley, 1922 Orndorff & Kline, 1924). Despite extensive investiga-tions with repect to its synthesis, there has not been a crys-tallographic study of (I). The present study reports the crystal structure of 2-(2,4-dihydroxybenzoyl)benzoic acid at room temperature.
Selected bond lengths and angles for (I) are given in Table 1. The 2,4-dihydroxybenzoyl and o-benzoic acid moieties are each essentially planar, with maximum deviations from each plane of 0.0170 (21) A˚ for C5 and 0.0417 (17) A˚ for O5, and the angle between these planes is 86.79 (4). In addition to an
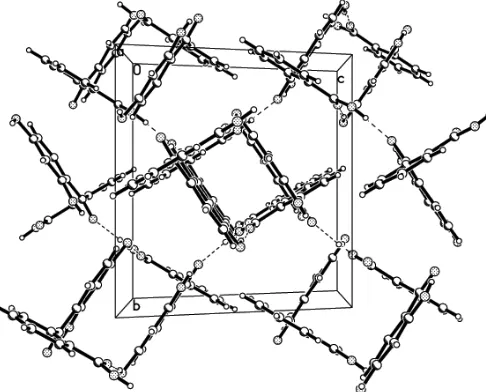
intramolecular O—H O hydrogen bond, intermolecular O—H O hydrogen bonds connect molecules to form a two-dimensional network parallel to (101) (see Table 2 and Fig. 2).
Experimental
The title compound was prepared according to the method described by Orndorff & Kline (1924). Crystals suitable for X-ray diffraction were obtained by slow evaporation of a solution in methanol and water.1H NMR (DMSO-d
6):6.21 (dd,
4J= 2.0 Hz,3J= 8.8 Hz, 1H),
6.32 (d, J= 2.4 Hz, 1H), 6.92 (d, J= 8.8 Hz, 1H), 7.36 (dd,4J= 0.8 Hz,
3J= 7.6 Hz, 1H), 7.58–7.70 (m, 2H), 8.08 (dd,4J= 0.8 Hz,3J= 8.0 Hz,
1H), 10.68 (s, 1H), 12.22 (s, 1H), 13.15 (s, 1H);13C NMR (DMSO-d6):
103.63, 109.03, 114.95, 128.63, 130.66, 131.48, 133.42, 136.06, 142.00, 166.59, 168.70, 202.82.
Crystal data
C14H10O5
Mr= 258.22
Monoclinic,P21=n
a= 10.331 (3) A˚
b= 11.628 (4) A˚
c= 11.640 (4) A˚
= 116.034 (5) V= 1256.4 (7) A˚3
Z= 4
Dx= 1.365 Mg m
3
MoKradiation Cell parameters from 2633
reflections
= 2.6–26.5
= 0.11 mm1
T= 294 (2) K Block, colourless 0.300.220.22 mm
Data collection
Bruker SMART CCD diffractometer
’and!scans
Absorption correction: multi-scan (SADABS; Bruker, 1997)
Tmin= 0.963,Tmax= 0.977
6755 measured reflections
2626 independent reflections 1762 reflections withI> 2(I)
Rint= 0.099 max= 26.7 h=12!12
k=13!14
l=14!13
Refinement
Refinement onF2
R[F2> 2(F2)] = 0.049
wR(F2) = 0.146
S= 1.07 2626 reflections 184 parameters
H atoms treated by a mixture of independent and constrained refinement
w= 1/[2(F
o2) + (0.0478P)2
+ 0.3516P]
whereP= (Fo2+ 2Fc2)/3
(/)max< 0.001
max= 0.27 e A˚3
min=0.21 e A˚3
Table 1
Selected geometric parameters (A˚ ,).
O2—C1 1.316 (2)
O4—C10 1.353 (3)
C1—C2 1.488 (3)
C3—C8 1.513 (3)
C8—C9 1.436 (3)
O1—C1—O2 122.6 (2) C4—C3—C8 116.49 (17) C2—C3—C8 124.45 (17)
O3—C8—C9 121.81 (17) C14—C9—C8 121.67 (17) C10—C9—C8 121.12 (18)
O1—C1—C2—C7 179.5 (2) O2—C1—C2—C7 0.0 (3) O1—C1—C2—C3 0.7 (3)
O2—C1—C2—C3 178.9 (2) O3—C8—C9—C14 178.77 (19) O3—C8—C9—C10 0.5 (3)
Table 2
Hydrogen-bonding geometry (A˚ ,).
D—H A D—H H A D A D—H A
O2—H2 O3i
0.89 (3) 1.76 (3) 2.643 (2) 177 (3) O4—H4 O3 0.90 (3) 1.80 (3) 2.602 (2) 148 (3) O5—H5 O1ii
0.89 (4) 1.89 (4) 2.764 (2) 169 (3)
Symmetry codes: (i)1 2þx;
1 2y;
1
2þz; (ii)x;1y;1z.
All H atoms bonded to C atoms were included in calculated positions, with C—H = 0.93 A˚ . They were included in the refinement in riding-model approximation, with Uiso(H) = 1.2Ueq(C). The H
atoms bonded to O atoms were refined independently with isotropic displacement parameters.
Data collection:SMART(Bruker, 1997); cell refinement:SAINT
(Bruker, 1997); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics:
SHELXTL (Bruker, 1997); software used to prepare material for publication:SHELXTL.
References
Baeyer (1876).Justus Liebigs Ann. Chem.183, 23–24. Bollmann, F. (1922).J. Prakt. Chem.104, 123–126.
Bruker (1997).SADABS,SMART,SAINTandSHELXTL(Version 5.10). Bruker AXS Inc., Madison, Wisconsin, USA.
Orndorff, W. R. & Adamson, W. A. (1918).J. Am. Chem. Soc.40, 1235–1257. Orndorff, W. R. & Kelley, L. (1922).J. Am. Chem. Soc.44, 1518–1527. Orndorff, W. R. & Kline, E. (1924).J. Am. Chem. Soc.46, 2276–2291.. Sheldrick, G. M. (1997). SHELXS97 and SHELXL97. University of
[image:2.610.312.568.72.196.2]Go¨ttingen, Germany. Figure 1
[image:2.610.319.562.251.447.2]A view of the molecular of (I). Displacement ellipsoids are drawn at the 30% probability level and H atoms are shown as small spheres of arbitrary radii.
Figure 2
supporting information
sup-1 Acta Cryst. (2005). E61, o1769–o1770
supporting information
Acta Cryst. (2005). E61, o1769–o1770 [https://doi.org/10.1107/S1600536805014844]
2-(2,4-Dihydroxybenzoyl)benzoic acid
Yang Li, Feng-Yan Ge, Li-Gong Chen, Chuan-Ming Dong, Xi-Long Yan, Er-Hong Duan, Tao Zeng,
Yue-Cheng Zhang and Guo-Yi Bai
2-(2,4-Dihydroxybenzoyl)benzoic acid
Crystal data
C14H10O5
Mr = 258.22
Monoclinic, P21/n
Hall symbol: -P 2yn a = 10.331 (3) Å b = 11.628 (4) Å c = 11.640 (4) Å β = 116.034 (5)° V = 1256.4 (7) Å3
Z = 4
F(000) = 536 Dx = 1.365 Mg m−3
Melting point: 474 K
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 2633 reflections θ = 2.6–26.5°
µ = 0.11 mm−1
T = 294 K Block, colourless 0.30 × 0.22 × 0.22 mm
Data collection
Bruker SMART CCD diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
φ and ω scans
Absorption correction: multi-scan (SADABS; Bruker, 1997) Tmin = 0.963, Tmax = 0.977
6755 measured reflections 2626 independent reflections 1762 reflections with I > 2σ(I) Rint = 0.099
θmax = 26.7°, θmin = 2.2°
h = −12→12 k = −13→14 l = −14→13
Refinement
Refinement on F2
Least-squares matrix: full R[F2 > 2σ(F2)] = 0.049
wR(F2) = 0.146
S = 1.07 2626 reflections 184 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H atoms treated by a mixture of independent and constrained refinement
w = 1/[σ2(F
o2) + (0.0478P)2 + 0.3516P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.27 e Å−3
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
O1 0.28344 (15) 0.31036 (16) 0.44868 (15) 0.0581 (5)
O2 0.51504 (16) 0.2925 (2) 0.49583 (19) 0.0746 (6)
H2 0.516 (3) 0.250 (3) 0.560 (3) 0.087 (10)*
O3 0.01604 (15) 0.33945 (15) 0.18108 (15) 0.0572 (5)
O4 −0.17529 (15) 0.41733 (17) 0.25016 (16) 0.0608 (5)
H4 −0.134 (3) 0.369 (3) 0.216 (3) 0.088 (10)*
O5 −0.0508 (2) 0.75316 (16) 0.5080 (2) 0.0746 (6)
H5 −0.132 (4) 0.733 (3) 0.511 (3) 0.103 (11)*
C1 0.3836 (2) 0.32926 (19) 0.42363 (19) 0.0426 (5)
C2 0.3694 (2) 0.39474 (18) 0.30884 (18) 0.0400 (5)
C3 0.23439 (19) 0.43702 (18) 0.22257 (17) 0.0388 (5)
C4 0.2224 (2) 0.4955 (2) 0.1141 (2) 0.0516 (6)
H4A 0.1331 0.5235 0.0563 0.062*
C5 0.3419 (2) 0.5127 (2) 0.0909 (2) 0.0547 (6)
H5A 0.3322 0.5515 0.0177 0.066*
C6 0.4747 (2) 0.4722 (2) 0.1764 (2) 0.0556 (6)
H6 0.5550 0.4846 0.1616 0.067*
C7 0.4888 (2) 0.4131 (2) 0.2845 (2) 0.0499 (6)
H7 0.5786 0.3853 0.3415 0.060*
C8 0.09718 (19) 0.42241 (18) 0.23772 (18) 0.0401 (5)
C9 0.05604 (18) 0.50795 (18) 0.30447 (18) 0.0393 (5)
C10 −0.07981 (19) 0.50272 (19) 0.30834 (19) 0.0433 (5)
C11 −0.1173 (2) 0.5859 (2) 0.3735 (2) 0.0498 (6)
H11 −0.2072 0.5832 0.3736 0.060*
C12 −0.0219 (2) 0.67262 (19) 0.4384 (2) 0.0517 (6)
C13 0.1129 (2) 0.6803 (2) 0.4358 (2) 0.0539 (6)
H13 0.1765 0.7395 0.4784 0.065*
C14 0.1485 (2) 0.59974 (19) 0.3700 (2) 0.0458 (5)
H14 0.2373 0.6053 0.3681 0.055*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
O1 0.0417 (8) 0.0787 (12) 0.0642 (10) 0.0093 (8) 0.0327 (7) 0.0198 (9)
O2 0.0337 (8) 0.1151 (17) 0.0712 (11) 0.0076 (9) 0.0195 (8) 0.0453 (12)
supporting information
sup-3 Acta Cryst. (2005). E61, o1769–o1770
O4 0.0333 (8) 0.0822 (13) 0.0688 (11) −0.0122 (8) 0.0241 (8) −0.0166 (10)
O5 0.0871 (13) 0.0554 (11) 0.1157 (17) −0.0002 (10) 0.0762 (13) −0.0117 (10)
C1 0.0318 (10) 0.0511 (13) 0.0438 (11) 0.0005 (9) 0.0157 (8) 0.0027 (10)
C2 0.0350 (10) 0.0454 (11) 0.0407 (10) −0.0005 (8) 0.0176 (8) 0.0007 (9)
C3 0.0331 (9) 0.0457 (11) 0.0399 (10) 0.0005 (8) 0.0182 (8) −0.0023 (9)
C4 0.0394 (10) 0.0645 (15) 0.0489 (12) 0.0053 (10) 0.0177 (9) 0.0109 (11)
C5 0.0543 (12) 0.0654 (15) 0.0511 (12) 0.0005 (12) 0.0294 (10) 0.0145 (12)
C6 0.0459 (12) 0.0698 (16) 0.0641 (14) −0.0043 (11) 0.0362 (11) 0.0060 (12)
C7 0.0332 (10) 0.0647 (15) 0.0547 (13) 0.0034 (10) 0.0221 (9) 0.0064 (11)
C8 0.0288 (9) 0.0530 (13) 0.0359 (10) −0.0024 (9) 0.0118 (8) −0.0012 (9)
C9 0.0283 (9) 0.0490 (12) 0.0397 (10) 0.0039 (8) 0.0141 (8) 0.0042 (9)
C10 0.0289 (9) 0.0562 (13) 0.0435 (11) 0.0019 (9) 0.0146 (8) 0.0057 (10)
C11 0.0367 (11) 0.0591 (14) 0.0619 (13) 0.0084 (10) 0.0295 (10) 0.0087 (12) C12 0.0570 (13) 0.0448 (12) 0.0676 (14) 0.0100 (11) 0.0406 (12) 0.0064 (11) C13 0.0525 (13) 0.0459 (12) 0.0738 (15) −0.0039 (10) 0.0374 (12) −0.0047 (11)
C14 0.0341 (10) 0.0505 (12) 0.0587 (13) 0.0003 (9) 0.0256 (9) 0.0017 (10)
Geometric parameters (Å, º)
O1—C1 1.212 (2) C5—C6 1.377 (3)
O2—C1 1.316 (2) C5—H5A 0.9300
O2—H2 0.89 (3) C6—C7 1.384 (3)
O3—C8 1.258 (2) C6—H6 0.9300
O4—C10 1.353 (3) C7—H7 0.9300
O4—H4 0.90 (3) C8—C9 1.436 (3)
O5—C12 1.355 (3) C9—C14 1.412 (3)
O5—H5 0.89 (4) C9—C10 1.425 (3)
C1—C2 1.488 (3) C10—C11 1.385 (3)
C2—C7 1.398 (3) C11—C12 1.381 (3)
C2—C3 1.402 (3) C11—H11 0.9300
C3—C4 1.391 (3) C12—C13 1.409 (3)
C3—C8 1.513 (3) C13—C14 1.359 (3)
C4—C5 1.389 (3) C13—H13 0.9300
C4—H4A 0.9300 C14—H14 0.9300
C1—O2—H2 110 (2) C2—C7—H7 119.7
C10—O4—H4 107 (2) O3—C8—C9 121.81 (17)
C12—O5—H5 107 (2) O3—C8—C3 117.90 (17)
O1—C1—O2 122.6 (2) C9—C8—C3 120.01 (17)
O1—C1—C2 123.41 (18) C14—C9—C10 117.20 (18)
O2—C1—C2 114.02 (17) C14—C9—C8 121.67 (17)
C7—C2—C3 119.36 (18) C10—C9—C8 121.12 (18)
C7—C2—C1 120.81 (18) O4—C10—C11 117.94 (17)
C3—C2—C1 119.83 (17) O4—C10—C9 121.81 (19)
C4—C3—C2 119.06 (17) C11—C10—C9 120.24 (19)
C4—C3—C8 116.49 (17) C12—C11—C10 120.34 (18)
C2—C3—C8 124.45 (17) C12—C11—H11 119.8
C5—C4—H4A 119.5 O5—C12—C11 122.7 (2)
C3—C4—H4A 119.5 O5—C12—C13 116.6 (2)
C6—C5—C4 119.9 (2) C11—C12—C13 120.7 (2)
C6—C5—H5A 120.1 C14—C13—C12 118.8 (2)
C4—C5—H5A 120.1 C14—C13—H13 120.6
C5—C6—C7 120.08 (19) C12—C13—H13 120.6
C5—C6—H6 120.0 C13—C14—C9 122.67 (19)
C7—C6—H6 120.0 C13—C14—H14 118.7
C6—C7—C2 120.66 (19) C9—C14—H14 118.7
C6—C7—H7 119.7
O1—C1—C2—C7 −179.5 (2) C2—C3—C8—C9 90.8 (3)
O2—C1—C2—C7 0.0 (3) O3—C8—C9—C14 178.77 (19)
O1—C1—C2—C3 −0.7 (3) C3—C8—C9—C14 −7.5 (3)
O2—C1—C2—C3 178.9 (2) O3—C8—C9—C10 −0.5 (3)
C7—C2—C3—C4 0.5 (3) C3—C8—C9—C10 173.21 (17)
C1—C2—C3—C4 −178.38 (19) C14—C9—C10—O4 −179.06 (18)
C7—C2—C3—C8 179.7 (2) C8—C9—C10—O4 0.3 (3)
C1—C2—C3—C8 0.8 (3) C14—C9—C10—C11 0.5 (3)
C2—C3—C4—C5 −0.3 (3) C8—C9—C10—C11 179.82 (19)
C8—C3—C4—C5 −179.5 (2) O4—C10—C11—C12 177.71 (19)
C3—C4—C5—C6 −0.5 (4) C9—C10—C11—C12 −1.9 (3)
C4—C5—C6—C7 0.9 (4) C10—C11—C12—O5 −177.2 (2)
C5—C6—C7—C2 −0.6 (4) C10—C11—C12—C13 2.1 (3)
C3—C2—C7—C6 −0.1 (3) O5—C12—C13—C14 178.3 (2)
C1—C2—C7—C6 178.8 (2) C11—C12—C13—C14 −1.0 (3)
C4—C3—C8—O3 84.0 (3) C12—C13—C14—C9 −0.4 (3)
C2—C3—C8—O3 −95.2 (2) C10—C9—C14—C13 0.6 (3)
C4—C3—C8—C9 −90.0 (2) C8—C9—C14—C13 −178.7 (2)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
O2—H2···O3i 0.89 (3) 1.76 (3) 2.643 (2) 177 (3)
O4—H4···O3 0.90 (3) 1.80 (3) 2.602 (2) 148 (3)
O5—H5···O1ii 0.89 (4) 1.89 (4) 2.764 (2) 169 (3)