Original Article
Over-expression of TOP2A as a prognostic biomarker
in patients with glioma
Tianmin Zhou
1, Yan Wang
2, Dongmeng Qian
1, Qing Liang
1, Bin Wang
11Key Laboratory of Medicine and Biotechnology of Qingdao, Department of Microbiology, Medical College of Qingdao University, Qingdao, Shandong, P. R. China; 2Department of Pathology, The Affiliated Hospital of Qingdao University, Qingdao, Shandong, P. R. China
Received December 7, 2017; Accepted January 19, 2018; Epub March 1, 2018; Published March 15, 2018
Abstract: Topoisomerase (DNA) II alpha (TOP2A), an enzyme that controls and alters the topologic states of DNA during transcription, is aberrantly expressed in many cancers. However, few studies have investigated expression of TOP2A and its clinical significance in glioma. We retrieved six independent investigations from the Oncomine da-tabase and found that TOP2A is highly expressed in glioma tissues compared with corresponding normal controls. Similar results were also found in clinical specimens at the protein level. Immunohistochemical analysis indicated that TOP2A over expression was highly correlated with grade stage, KI67 positive percentage, IDH1 mutation, and age, but other clinical parameters such as sex distribution and tumor size were barely associated with high TOP2A gene expression. Meanwhile we used Prognos can to assess the prognostic value of TOP2A expression in glioma patients, and found that high expression was associated with poor prognosis of patients with glioma. Furthermore, we used the Gene-Cloud of Biotechnology Information (GCBI) bioinformatics platform predict the role of TOP2A in glioma. It was not only involved in DNA replication, chromosome condensation, and responses to DNA damage stimuli, but also promoted cancer cell mitotic cell cycle and apoptosis, and phosphatidylinositol-mediated signaling by regulating gene expression. By these approaches we demonstrate that TOP2A may be a reliable prognostic factor or therapeutic target in glioma.
Keywords: TOP2A, glioma, marker, prognosis
Introduction
Glioma remains the most common adult brain
tumor which is slightly more common in men
than in women [1], with 7.3 cases pr. 100,000
person-years. High-grade gliomas are present
in 85% and low-grade glioma in 15% with 5-
year overall survival of 82, 54, 22 and 3% for
grade I, II, III and IV, respectively [2]. Within a
five year follow-up time, most patients had a
recurrent tumor and died of the disease [3].
Despite much effort focused on improving the
diagnosis and therapy of glioma, the clinical
outcome of glioma is still unsatisfactory [4,
5]. Consequently, more studies are needed to
explore the potential mechanism and detect
effective diagnostic and prognostic biomark-
ers to prolong the lives of glioma patients.
TOP2A gene encodes a DNA topoisomerase
that controls and alters the topologic state of
DNA during transcription. This nuclear enzyme
is involved in processes such as chromosome
condensation, chromatid separation, and the
relief of torsional stress that occurs during
DNA transcription and replication [6]. Its
aber-rant expression has been associated with mu-
ltiple tumor prognostics [7], including lung
cancer, breast cancer, pancreatic
adenocarci-noma, and colorectal cancer [8-11]. There
are few investigations analyzing expression of
TOP2A in glioma [12], and due to the limited
number of studies and few clinical samples, no
role has been detected.
Materials and methods
Oncomine analysis
The Oncomine database (http://www.oncomi-
ne.org) incorporates 264 independent
datas-ets including 35 cancer types and supports
various methods of analysis, including
molecu-lar concepts, interactome, and meta-analysis
[13]. Thus, we detected the level of TOP2A
in different cancers including hematological
malignancies and solid tumors and adopted
the Oncomine database online tools meta-an-
alysis to estimate the expression of TOP2A in
glioma and normal brain control that come
from 6 different datasets.
PrognoScan
PrognoScan (http://www.prognoscan.org) is a
comprehensive platform for evaluating pot-
ential tumor biomarkers and therapeutic
tar-gets. Based on a large collection of cancer
microarray datasets with clinical annotation,
PrognoScan is a useful online tool for
assess-ing the association between specific gene ex-
pression and prognosis in patients with can-
cer [14]. We used the PrognoScan platform to
validate the prognostic value of TOP2A expr-
ession in patients with glioma.
Patients and specimens
Forty-six glioma tumor tissue samples were
collected from the Affiliated Hospital of
Qing-dao University at QingQing-dao in China from July
2014 to December 2016. All the patients we-
re with tumors suitable for resection accord-
ing to the Chinese guideline of surgical
treat-ment strategy for glioma in 2012, and among
them, 16 cases were in gradeII, 9 cases were
grade III, and 21 cases were glioma. Altoge-
ther, 46 cases of formalin-fixed paraffin-em-
bedded (FFPE) specimens were used for Im-
munohistochemistry.
In this study, the inclusion criteria were as
the following: (1) Pathological diagnosis of gli-
oma tumors; (2) A diagnosis that was cons-
istent with the histological diagnostic criteria
of the World Health Organization [15]; (3) Tu-
mor is the primary tumor in the brain, and pa-
tients did not receive pre-operative anti-can-
cer treatment, and there were no extrahepa-
tic metastases; (4) For the use of archived
tis-sue specimens from these patients,
permis-sion from the Affiliated Hospital of Qingdao
University was granted. (5) A representative
sample was taken from each FFPE tissue and
hematoxylin and eosin (HE) staining was
per-formed. Results were reviewed by two
experi-enced pathologists.
Immunohistochemical staining and evaluation
A traditional immunohistochemical (IHC)
stain-ing protocol was used in this study. Briefly,
tis-sue slices was deparaffinized in xylene and
rehydrated with different concentrations of et-
hanol alcohol. Then the section was treated
with 3% hydrogen peroxide, followed by anti-
gen retrieval with 10 mM citrate buffer (pH
6.0) with microwave. After being blocked with
10% goat serum for 30 min, incubation with
the primary antibody was done overnight at
4°C with anti-TOP2A antibody (mouse
onoclo-nal antibody, 1:100 dilution, ZM-0245, Zhong-
shanjinqiao, China) overnight at 4°C, followed
by a peroxidase-labeled secondary antibody.
Immunoreaction score distribution (IOD)
mea-surement, as reported previously, was used
to determine the positive-staining density [16].
A Leica CCD camera DFC420 connected to a
Leica DM IRE2 microscope (Leica Microsyst-
ems Imaging Solutions, Ltd., Cambridge, UK)
was used as an imaging system. High-power
magnification (×40) was used to photograph
representative fields. The IODs in each image
were measured and counted using Image-Pro
Plus v6.0 software (Media Cybernetics, Inc.,
Bethesda, MD, USA).
X-tile analysis
TOP2A expression was assessed with X-tile
plots. TOP2A expression was expressed as
IOD and the optimization of cutoff points was
based on outcomes. The cutoff score derived
from 46 cases of training set by a standard
log-rank method was used to assess statisti-
cal significance.
P
values were obtained from
a lookup table.
Gene-cloud of biotechnology information
(GCBI)
bioinformat-ics. It creates a “gene knowledge
base” which encompasses biolo-
gy, medicine, informatics, comput-
er science, mathematics, graphics,
and other disciplines. GCBI includ-
es more than 120 million copies
of genomic samples, approximately
90,000 copies of tumor samples
and more than 17 million copies of
genetic information. Therefore, GC-
BI is a good bioinformatics analys-
is platform and has provided data
analysis support for many studies
on cancer research [17-20]. In this
study, we used GCBI to identify
dif-f-ferentially expressed genes (DEGs)
between glioma tissue and normal
brain control and finally predict the
role of TOP2A in gliomas.
Statistical analysis
[image:3.612.93.365.67.720.2]Figure 2. TOP2A expression levels in peri-tumoral tis-sues (A) and glioma (B) (×40). (C) Box plot indicating the mean staining intensity of paired glioma and peri-tumoral tissues. (D) Paired peri-peri-tumoral tissue and glioma tissue in the same section (D) (×10).
Table 1.
Meta-analysis of TO2PA gene expression from six Oncomine databases
Datasets (sample size) Comparison groups ChangeFold P-value Overexpression Gene Rank Sun Brain (180) Glioblastoma vs. Normal 102.544 2.37E-27 22 (in top 1%)
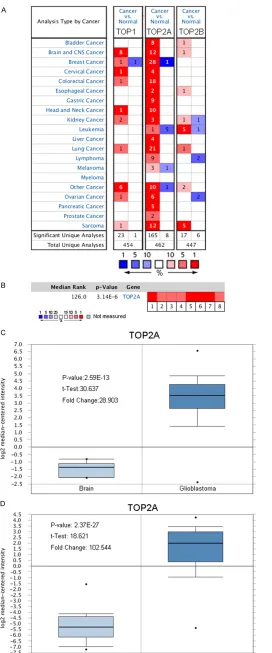
Oligodendroglioma vs. Normal 28.449 8.4E-18 27 (in top 1%) Anaplastic Astocytoma 34.640 4.10E-12 27 (in top 1%) Liang Brain (33) Glioblastoma vs. Normal 2.202 6.27E-6 115 (in top 2%) TCGA Brain (552) Glioblastoma vs. Normal 28.903 2.59E-13 115 (in top 2%) French Brain Statistics (29) Anaplastic Oligodendroglioma VS. Brain 9.685 1.15E-8 115 (in top 2%) Shai Brain Statistics (34) Glioblastoma vs. White Matter 2.291 1.17E-6 227 (in top 3%) Murat Brain Statistics (84) Glioblastoma vs. Normal 13.755 2.17E-11 262 (in top 2%) Note: A meta-analysis of TO2PA gene expression from six Oncomine databases where colored squares indicate the median rank for TOP2A (vs. Normal tissue) across 8 analyses. French Brain (1), Liang Brain (2), Murat Brain (3), Shai Brain (4), Sun Brain (5-7) and TCGA (8). The P value is given for the medianrank analysis.
Variance. Survival curves for TOP2A expres-
sion were generated using the Kaplan-Meier
method and compared using the log-rank test.
P
values < 0.05 were considered statistically
significant. Each statistical analysis of online
database was completed by usedits online tool.
Results
TOP2A is distinctively overexpressed in glioma
than normal brain
Table 2.
Association of TOP2A expression with
pathological characteristics of glioma (46
cases)
Pathological
category numberCase
TOP2A
P-value Low High
Sex Male 29 11 18 0.1264 Female 17 11 6
Age ≥ 50 23 5 18 0.0009 < 50 23 17 16
Tumor size ≥ 3 cm 21 10 11 1.0000 < 3 cm 25 12 13
[image:5.612.91.293.517.652.2]IDH1 IDH1 (+) 15 12 3 0.0113 IDH1 (-) 16 5 11
Figure 3. Correlation analysis of TOP2A with clinicopathological features of glioma. A, B: Expression of TOP2A was significantly associated with grade stage. C: KI67 positive percentage.
normal tissues in a dataset with 552 samples
that derived from TCGA database [21] (
Figure
1C
). In another dataset from Sun L study [22],
TOP2A was 102.544 fold elevated in glioma
samples as compared with
normal tissues (P =
2.37*10-27) (
Figure 1D
).
Elevation of TOP2A in glioma
compared with peri-tumoral
tissues by immunochemistry
We also analyzed the
expres-sion levels of TOP2A protein in
46 glioma tumors and paired
peri-tumoral tissues by
immu-nochemistry (IHC). Expression
of TOP2A protein levels was
frequently higher in the glioma
tumor tissues (
Figure 2A
) than
in peri-tumoral tissues (
Figu-
re 2B
). These differences we-
re statistically significant (
Fi-
gure 2C
; P < 0.0001, paired t
test). Meanwhile we found th-
at expression of TOP2A is po-
sitive in all samples of glioma.
In 43 cases, glioma tissues
and peri-tumoral tissue were
located in one area, and the
picture clearly revealed that
TOP2A expression was higher
in peri-tumoral tissue than gli-
oma tissue (
Figure 2D
).
Association of TOP2A
expres-sion with clinicopathological
features of glioma patients
As show in
Figure 3A
and
3B
, Immunoreaction
score distribution (IOD) of TOP2A significant-
ly increased along with grad stage of glioma (F
= 11.91, P < 0.0001) and KI67 positive
per-centage (Spearman’s r = 0.7788, P < 0.0001).
We also learned the association of the expr-
ession of TOP2A with other clinicopathological
features in glioma, as shown in
Table 2
. Ex-
pression of TOP2A association correlated wi-
th IDH1 mutation (P = 0.0113) and age (P =
0.0009), but other clinical parameters, such as
sex distribution (P = 0.1264) and tumor size (P
= 1.0000) were barely associated with high
TOP2A gene expression.
High mRNA expression of TOP2A is an
unfavor-able prognostic factor for glioma
2925 (P < 0.01, Flod change > 2) significant
DEGs were identified, of which 1041
over-pre-sented and 1884 showed an attenuated be-
havior (
Figure 5B
and
5C
). We found that TOP-
2A was significantly overexpressed in glioma
samples (Fold Change = 6.447, t = 17.9485, P
< 0.01). We then performed a Gene Ontology
analysis of 2925 differentially expressed ge-
nes (
Figure
5D
). Fortunately, we found that
TOP2A in the enriched set was associated wi-
th mitotic cell cycle, positive regulation of tr-
anscription from RNA polymerase II promoter,
positive regulation of apoptotic process, DNA
replication, phosphatidylinositol-mediated
sig-naling, response to DNA damage stimulus,
chromosome condensation. So we speculate
that the elevation of TOP2A not only takes
part in DNA replication, chromosome
conden-sation, and response to DNA damage stimu-
lus, but also associates with mitotic cell cycle,
apoptotic processes, and
phosphatidylinositol-mediated signaling.
Discussion
Glioma, a tumor of central nervous system,
which mainly develops from the macroglial
cells, presents the highest prevalence and mo-
rtality risk. The difficult treatment of patients
with glioma is primarily attributed to recurren-
ce and resistance to chemoradiotherapy
[29-31]. Despite the challenges of treatment, it is
rewarding to illustrate the pathogenesis of gli-
oma, as well as to develop novel prognostic
strategies and discover effective therapeutic
approaches. TOP2A is a nuclear enzyme that
mainly involved in processes such as chro-
mosome condensation, chromatid separation,
and the relief of torsional stress that occurs
during DNA transcription and replication [6]. It
has been reported that TOP2A is a sensitive
and specific marker in actively proliferating
cells (in the late S, G2, and M phases of the cell
cycle) which suggests a role in a wide range
of human cancers [32, 33].
[image:6.612.89.288.71.315.2]In the present study, through the Oncomine
databases, we found that TOP2A was
signifi-cantly over expressed in glioma compared with
normal brain samples. Similar results were
found in 46 gliomas and paired peri-tumoral
tissues by immunochemistry byanalysis of the
association of TOP2A expression with
clinico-pathological features of glioma patients. TOP-
2A significantly increased along with grade
Figure 4. Kaplan-Meier survival curves generated from PrognoScan for TOP2A mRNA expression in tients with glioma. A: Overall survival curves for pa-tients with glioma in the GEO dataset GSE4271. B: Patients with glioma in the GEO dataset GSE4412. HR = hazard ratio.
cordingly, we hypothesized that TOP2A expr-
ession may be a prognostic factor for patients
with glioma. We used the PrognoScan
data-base to analyze the correlation of TOP2A ex-
pression with overall survival in GSE4271 and
GSE4412 enrolled from the Gene Expression
Omnibus (GEO) datasets with a total of 153
cases. The results show that TOP2A expres-
sion is highly associated with overall survival
of glioma patients (log-rank test, P < 0.001,
P = 0.0016,
Figure 4A
and
4B
).
Prediction of the role of TOP2A in glioma
stage of glioma, KI67 positive percentage,
IDH1 mutation and age, but other clinical
parameters, such as sex distribution and tu-
mor size were barely associated with high
TOP2A gene expression. However, there was
no evidence revealing arole of highly express-
ed TOP2A in the prognosis of glioma. In our
study, according to the PrognoScan database,
we could find its prognostic value: high expr-
ession of TOP2A caused shorter overall survi-
val time and shorter disease-free survival time.
Thus, the expression of TOP2A might become
a potential biomarker for the prognosis of
glioma.
As for the functional and pathway enrichment
analysis, TOP2A was strongly emphasized in
cell cycle pathways and DNA replication
pro-cesses, which areconnected with its physiolo-
gical functions [34, 35]. In our study, GCBI
anal-ysis found that mitotic cell cycle, positive
regu-lation of transcription from RNA polymerase II
promoter, positive regulation of apoptotic
pro-cess, DNA replication, phosphatidylinositol-me-
diated signaling, response to DNA damage
stimulus, and chromosome condensation pa-
thways were highly enriched in glioma sampl-
es with TOP2A up-regulated. Furthermore,
ear-lier clinical data indicate that TOP2A mRNA
and protein expression might participate in the
process of chromosome condensation,
chro-matid separation, and the relief of torsional
stress that occurs during DNA transcription and
replication [6]. According to our analysis, we
speculate that high expression of TOP2A at
mRNA and protein levels might be one of the
causes of cell cycle apoptotic process and
phosphatidylinositol-mediated signaling.
In conclusion, we used bioinformatics and
immunochemistry analysis to define the expr-
ession level of TOP2A in glioma. TOP2A was
identified in association with the progression
and prognosis of glioma, probably regulating
cell cycle apoptotic processes and
phosphati-dylinositol-mediated signaling.
Disclosure of conflict of interest
None.
Address correspondence to: Tianmin Zhou, Key Laboratory of Medicine and Biotechnology of Qing- dao, Department of Microbiology, Medical College of Qingdao University, 308 Ningxia Road, Qingdao, P.
R. China. E-mail: 329637042@163.com; Bin Wang, School of Basic Medical Sciences, Qingdao Uni- versity, Qingdao, P. R. China. Tel: +86-131-0772-05- 70; E-mail: wangbinqindao@163.com
References
Sumer SO, Sun Y, Tasman N, Taylor BS, Voet D, Weinhold N, Weinstein JN, Yang D, Yoshihara K, Zheng S, Zhang W, Zou L, Abel T, Sadeghi S, Cohen ML, Eschbacher J, Hattab EM, Raghu-nathan A, Schniederjan MJ, Aziz D, Barnett G, Barrett W, Bigner DD, Boice L, Brewer C, Cala-tozzolo C, Campos B, Carlotti CG Jr, Chan TA, Cuppini L, Curley E, Cuzzubbo S, Devine K, DiMeco F, Duell R, Elder JB, Fehrenbach A, Fin-occhiaro G, Friedman W, Fulop J, Gardner J, Hermes B, Herold-Mende C, Jungk C, Kendler A, Lehman NL, Lipp E, Liu O, Mandt R, McGraw M, Mclendon R, McPherson C, Neder L, Nguy-en P, Noss A, Nunziata R, Ostrom QT, Palmer C, Perin A, Pollo B, Potapov A, Potapova O, Rath-mell WK, Rotin D, Scarpace L, Schilero C, Sen-ecal K, Shimmel K, Shurkhay V, Sifri S, Singh R, Sloan AE, Smolenski K, Staugaitis SM, Steele R, Thorne L, Tirapelli DP, Unterberg A, Valluru-palli M, Wang Y, Warnick R, Williams F, Wolin-sky Y, Bell S, Rosenberg M, Stewart C, Huang F, Grimsby JL, Radenbaugh AJ, Zhang J. Compre-hensive, integrative genomic analysis of dif-fuse lower-grade gliomas. N Engl J Med 2015; 372: 2481-98.
[2] Rasmussen BK, Hansen S, Laursen RJ, Kostel-janetz M, Schultz H, Nørgård BM, Guldberg R and Gradel KO. Epidemiology of glioma: clini-cal characteristics, symptoms, and predictors of glioma patients grade I-IV in the the Danish neuro-oncology registry. J Neurooncol 2017; 135: 571-579.
[3] Wion D. Therapeutic dormancy to delay post-surgical glioma recurrence: the past, present and promise of focal hypothermia. J Neuroon-col 2017; 133: 447-454.
[4] van den Bent M, Chinot OL and Cairncross JG. Recent developments in the molecular charac-terization and treatment of oligodendroglial tumors. Neuro Oncol 2003; 5: 128-38. [5] Kalpathy-Cramer J, Gerstner ER, Emblem KE,
Andronesi O and Rosen B. Advanced magnetic resonance imaging of the physical processes in human glioblastoma. Cancer Res 2014; 74: 4622-4637.
[6] Tsavaris N, Lazaris A, Kosmas C, Gouveris P, Kavantzas N, Kopterides P, Papathomas T, Ara-pogiannis G, Zorzos H, Kyriakou V and Pat-souris E. Topoisomerase I and IIα protein ex-pression in primary colorectal cancer and recurrences following 5-fluorouracil-based ad-juvant chemotherapy. Cancer Chemother Pharmacol 2009; 64: 391-398.
[7] Heestand GM, Schwaederle M, Gatalica Z, Ar-guello D and Kurzrock R. Topoisomerase ex-pression and amplification in solid tumours: Analysis of 24,262 patients. Eur J Cancer 2017; 83: 80-7.
[8] Wang TL, Ren YW, Wang HT, Yu H and Zhao YX. Association of topoisomerase II (TOP2A) and dual-specificity phosphatase 6 (DUSP6) single nucleotide polymorphisms with radiation treat-ment response and prognosis of lung cancer in Han Chinese. Med Sci Monit 2017; 23: 984-993.
[9] Dimasgonzález J, Maldonadolagunas V, Díazchávez J, Lópezarellano ME, Muñozcama-cho J, Teránporcayo MA and Lagunasmartínez A. Overexpression of p53 protein is a marker of poor prognosis in Mexican women with breast cancer. Oncol Rep 2017; 37: 3026-3036. [10] Zhou Z, Liu S, Zhang M, Zhou R, Liu J, Chang Y
and Zhao Q. Overexpression of topoisomerase 2-alpha confers a poor prognosis in pancreatic adenocarcinoma identified by co-expression analysis. Dig Dis Sci 2017; 62: 2790-2800. [11] Yang FD and Jia ZL. [Expression of
topoisomer-ase II alpha in human colorectal carcinoma and its significance]. Nan Fang Yi Ke Da Xue Xue Bao 2010; 30: 1959-1961, 1964. [12] Arivazhagan A, Kumar DM, Sagar V, Patric IR,
Sridevi S, Thota B, Srividya MR, Prasanna K, Thennarasu K, Mondal N, Hegde AS, Chandra-mouli BA, Santosh V, Rao MR, Kondaiah P and Somasundaram K. Higher topoisomerase 2 al-pha gene transcript levels predict better prog-nosis in GBM patients receiving temozolomide chemotherapy: identification of temozolomide as a TOP2A inhibitor. J Neurooncol 2012; 107: 289-297.
[13] Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW and Klei-hues P. The 2007 WHO classification of tu-mours of the central nervous system. Acta Neuropathol 2007; 114: 97-109.
[14] Mizuno H, Kitada K, Nakai K and Sarai A. Prog-noScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Ge-nomics 2009; 2: 18.
[15] Zaczek AJ, Markiewicz A, Seroczynska B, Skokowski J, Jaskiewicz J, Pienkowski T, Olsze-wski WP, Szade J, Rhone P and Welnicka-Jaskiewicz M. Prognostic significance of TOP2A gene dosage in HER-2-negative breast cancer. Oncologist 2012; 17: 1246-1255.
[16] Franceschini A, Lin J, Mering CV and Jensen LJ. SVD-phy: improved prediction of protein func-tional associations through singular value de-composition of phylogenetic profiles. Bioinfor-matics 2016; 32: 1085-7.
[17] Feng A, Tu Z and Yin B. The effect of HMGB1 on the clinicopathological and prognostic features of non-small cell lung cancer. Oncotarget 2016; 7: 20507-20519.
mRNA-lncRNA signature with predictive and prognostic value. Cancer Res 2016; 76: 2105-14.
[19] Kong F, Wei H, Kai Z, Xiao W, Kou Y, You H, Zheng K and Tang R. Hepatitis B virus X protein promotes interleukin-7 receptor expression via NF-κB and Notch1 pathway to facilitate prolif-eration and migration of hepatitis B virus-relat-ed hepatoma cells. J Exp Clin Cancer Res 2016; 35: 172.
[20] Wang G, Cao R, Wang Y, Qian G, Dan HC, Jiang W, Ju L, Wu M, Xiao Y and Wang X. Simvastatin induces cell cycle arrest and inhibits prolifera-tion of bladder cancer cells via PPARγ signal-ling pathway. Sci Rep 2016; 6: 35783. [21]
https://tcga-data.nci.nih.gov/docs/publica-tions/tcga/.
[22] Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T and Fine HA. Neuronal and glioma-derived stem cell fac-tor induces angiogenesis within the brain. Can-cer Cell 2006; 9: 287-300.
[23] Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO and Israel MA. Gene ex-pression profiling reveals molecularly and clini-cally distinct subtypes of glioblastoma multi-forme. Proc Natl Acad Sci U S A 2005; 102: 5814-9.
[24] French PJ, Swagemakers SM, Nagel JH, Kou-wenhoven MC, Brouwer E, van der Spek P, Lu-ider TM, Kros JM, van den Bent MJ and Sillevis Smitt PA. Gene expression profiles associated with treatment response in oligodendroglio-mas. Cancer Res 2005; 65: 11335-11344. [25] Shai R, Shi T, Kremen TJ, Horvath S, Liau LM,
Cloughesy TF, Mischel PS and Nelson SF. Gene expression profiling identifies molecular sub-types of gliomas. Oncogene 2003; 22: 4918-4923.
[26] Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, Kouwenhoven MC, Hain-fellner JA, Heppner FL, Dietrich PY, Zimmer Y, Cairncross JG, Janzer RC, Domany E, Delorenzi M, Stupp R and Hegi ME. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradio-therapy in glioblastoma. J Clin Oncol 2008; 26: 3015-3024.
[27] Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transfor-mation and progression. Proc Natl Acad Sci U S A 2004; 101: 9309-9314.
[28] Kapur K, Xing Y, Ouyang Z and Wong WH. Exon arrays provide accurate assessments of gene expression. Genome Biol 2007; 8: R82. [29] Fack F, Espedal H, Keunen O, Golebiewska A,
Obad N, Harter PN, Mittelbronn M, Bähr O, Weyerbrock A, Stuhr L, Miletic H, Sakariassen PØ, Stieber D, Rygh CB, Lund-Johansen M, Zheng L, Gottlieb E, Niclou SP and Bjerkvig R. Bevacizumab treatment induces metabolic ad-aptation toward anaerobic metabolism in glio-blastomas. Acta Neuropathol 2015; 129: 115-31.
[30] Chen R, Cohen AL and Colman H. Targeted therapeutics in patients with high-grade glio-mas: past, present, and future. Curr Treat Op-tions Oncol 2016; 17: 42.
[31] Grosu AL, Weber WA, Franz M, Stärk S, Piert M, Thamm R, Gumprecht H, Schwaiger M, Molls M and Nieder C. Reirradiation of recurrent high-grade gliomas using amino acid PET (SPECT)/CT/MRI image fusion to determine gross tumor volume for stereotactic fractionat-ed radiotherapy. Int J Radiat Oncol Biol Phys 2005; 63: 511-519.
[32] Ravasz E, Somera AL, Mongru DA, Oltvai ZN and Barabási AL. Hierarchical organization of modularity in metabolic networks. Science 2002; 297: 1551-5.
[33] Lazaris AC, Kavantzas NG, Zorzos HS, Tsavaris NV and Davaris PS. Markers of drug resistance in relapsing colon cancer. J Cancer Res Clin Oncol 2002; 128: 114-118.
[34] Deweese JE and Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res 2009; 37: 738-48. [35] Xie ZC, Dang YW, Wei DM, Chen P, Tang RX,