metal-organic papers
m1292
Zhou and Yang [MnCl(C16H15N5)2]Cl3CH4O doi:10.1107/S1600536805017678 Acta Cryst.(2005). E61, m1292–m1294 Acta Crystallographica Section E
Structure Reports
Online
ISSN 1600-5368
Bis[bis(benzimidazol-2-ylmethyl)amine]-j
3N
,
N
000,
N
000000;
j
2N
,
N
000-chloromanganese(II)
chloride methanol trisolvate
Qing-Hua Zhou and Pin Yang*
Institute of Molecular Science, Chemical Biology and Molecular Engineering Laboratory of Education Ministry, Shanxi University, Taiyuan, Shanxi 030006, People’s Republic of China
Correspondence e-mail: yangpin@sxu.edu.cn
Key indicators
Single-crystal X-ray study
T= 203 K
Mean(C–C) = 0.006 A˚
Rfactor = 0.049
wRfactor = 0.071
Data-to-parameter ratio = 14.0
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
#2005 International Union of Crystallography Printed in Great Britain – all rights reserved
In the title compound, [MnClL2]Cl3CH4O, where L = bis(benzimidazol-2-ylmethyl)amine (C16H15N5), the coordina-tion geometry around the manganese(II) ion can be described as distorted octahedral, involving three benzimidazole N atoms, two tertiary amine atoms and a chloride anion. – Stacking interactions are found in two neighboring non-coordinated benzimidazole groups.
Comment
The imidazole of the histidine unit is one of the most common binding sites in metalloenzymes, such as alkaline phosphatase and superoxide dismutase (SOD). To understand the chem-istry of the assembly and catalytic mechanisms of these metalloenzymes it is logical to prepare potential model compounds by utilizing ligands incorporating the imidazole group. Thus the title MnIIcomplex, (I), with bis(benzimidazol-2-ylmethyl)amine (L) has been synthesized and its crystal structure is presented here.
The crystal structure of (I) consists of MnIIcomplex cations, chloride anions and methanol solvent molecules. The structure of the MnIIcomplex cation is depicted in Fig. 1. The MnIIion is surrounded by two L ligands and a chloride anion. One L ligand acts in a tridentate manner and the other acts in a bidentate manner. One chloride anion coordinates to the MnII ion to complete the distorted octahedral geometry. This structure is similar to that observed in another MnIIcomplex (Liaoet al., 2001). The Mn—Nimidazolebond lengths are shorter than the Mn—Nimino bonds (Table 1), in agreement with values reported previously (Yan et al., 2004). The Mn—Cl2 bond distance is also comparable to those reported for related
structures [2.316 (2)–2.538 (2) A˚ ; Oki et al., 1995]. The benzimidazole rings are planar, the maximum atomic devia-tion from the least-squares plane being less than 0.02 A˚ .
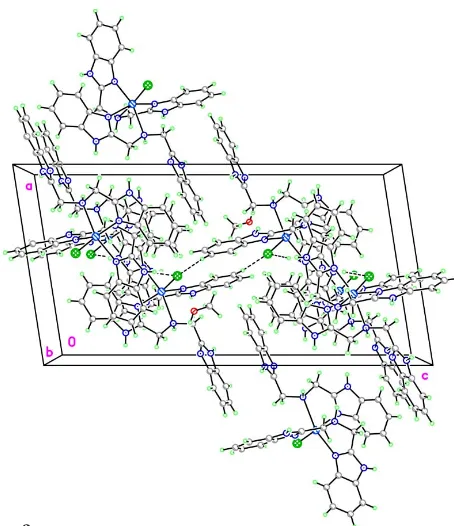
There is extensive hydrogen bonding in the crystal structure of (I) (Table 2). Neighboring uncoordinated benzimidazole groups, related by an inversion center, are parallel in an offset
fashion and separated by a face-to-face distance of
3.500 (6) A˚ . This fact indicates the existence of–stacking interactions, which stabilize the crystal packing together with the hydrogen-bonding interactions.
Experimental
All chemicals were of reagent grade and commercially available and were used without further purification.
Bis(benzimidazol-2-ylmeth-yl)amine (L) was prepared by a previously reported method (Berends & Stephan, 1984). LigandL(2.0 mmol) and MnCl24H2O (1.0 mmol) were dissolved in a methanol solution at room tempera-ture. Single crystals of (I) were obtained after three weeks. The elemental analysis agrees with the chemical composition of (I).
Crystal data
[MnCl(C16H15N5)2]Cl3CH4O Mr= 776.63
Monoclinic,P21=c a= 13.161 (3) A˚
b= 11.905 (3) A˚
c= 25.144 (7) A˚
= 98.794 (4)
V= 3893.2 (17) A˚3 Z= 4
Dx= 1.325 Mg m 3
MoKradiation Cell parameters from 2271
reflections
= 2.3–20.7
= 0.52 mm1 T= 203 (2) K Block, colorless 0.400.300.10 mm
Data collection
Bruker SMART CCD area-detector diffractometer
’and!scans
Absorption correction: multi-scan (SADABS; Bruker, 1998)
Tmin= 0.818,Tmax= 0.950
15680 measured reflections
6860 independent reflections 3596 reflections withI> 2(I)
Rint= 0.051 max= 25.0 h=15!7
k=14!14
l=29!29
Refinement
Refinement onF2 R[F2> 2(F2)] = 0.049 wR(F2) = 0.071 S= 0.93 6860 reflections 490 parameters
H atoms treated by a mixture of independent and constrained refinement
w= 1/[2
(Fo2) + (0.0102P)2]
whereP= (Fo2+ 2Fc2)/3
(/)max= 0.001
max= 0.48 e A˚
3
min=0.36 e A˚
3
Table 1
Selected geometric parameters (A˚ ,).
Mn—Cl2 2.425 (1)
Mn—N1 2.444 (3)
Mn—N2 2.187 (3)
Mn—N4 2.202 (2)
Mn—N6 2.389 (3)
Mn—N7 2.204 (3)
N2—Mn—N4 95.50 (9)
N2—Mn—N7 154.4 (1)
N4—Mn—N7 94.9 (1)
N2—Mn—N6 88.7 (1)
N4—Mn—N6 161.39 (11)
N7—Mn—N6 74.5 (1)
N2—Mn—Cl2 101.73 (8)
N4—Mn—Cl2 100.79 (8)
N7—Mn—Cl2 99.22 (7)
N6—Mn—Cl2 96.06 (8)
N2—Mn—N1 74.1 (1)
N4—Mn—N1 72.5 (1)
N7—Mn—N1 86.9 (1)
N6—Mn—N1 91.4 (1)
[image:2.610.46.295.74.239.2]Cl2—Mn—N1 171.39 (8)
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
O1—H1 O2 0.82 1.82 2.627 (4) 167
O2—H2A N9 0.82 1.93 2.698 (4) 155
O3—H3A Cl1i
0.82 2.34 3.114 (4) 158
N1—H1N Cl1i
0.93 (2) 2.54 (2) 3.247 (3) 133 (2) N3—H3N O1ii
0.89 (1) 1.88 (1) 2.724 (4) 158 (3) N5—H5N Cl2i
0.90 (1) 2.26 (1) 3.155 (3) 171 (3) N6—H6N Cl1i 0.92 (3) 2.41 (3) 3.292 (3) 161 (3) N8—H8N Cl1iii
0.89 (1) 2.21 (1) 3.096 (3) 178 (3) N10—H10N O3 0.90 (1) 1.91 (2) 2.754 (4) 156 (3)
C4—H4 Cl2iv 0.93 2.67 3.603 (4) 174
Symmetry codes: (i) xþ1;yþ1
2;zþ32; (ii) x;yþ1;z; (iii) xþ1;y;z; (iv) xþ1;yþ1;zþ1.
metal-organic papers
Acta Cryst.(2005). E61, m1292–m1294 Zhou and Yang [MnCl(C
[image:2.610.61.288.282.545.2]16H15N5)2]Cl3CH4O
m1293
Figure 2The packing of (I), showing the hydrogen bonding (dashed lines). Figure 1
[image:2.610.316.564.622.724.2]The hydroxy H atoms and methyl H atoms were placed in calcu-lated positions, with O—H = 0.82 and C—H = 0.96 A˚ , and refined as rigid groups rotated to fit the electron density, with Uiso(H) = 1.5Ueq(carrier). H atoms on N atoms were located in a difference Fourier map and isotropically refined. Other H atoms were placed in geometrically idealized positions, with C—H = 0.93 A˚ (aromatic) or 0.97 A˚ (methylene), and refined as riding with Uiso(H) = 1.2Ueq(carrier).
Data collection:SMART(Bruker, 1998); cell refinement:SAINT
(Bruker, 1998); data reduction: SAINT; program(s) used to solve structure: SHELXS97(Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics:
SHELXTL (Bruker, 1998); software used to prepare material for publication:SHELXTL.
This work was supported financially by the National Natural Science Foundation of China (grant No. 20171031).
References
Berends, H. P. & Stephan, D. W. (1984).Inorg. Chim. Acta,93, 173–178. Bruker (1998). SMART, SAINT, SHELXTL and SADABS (Version 5).
Bruker AXS Inc., Madison, Wisconsin, USA.
Liao, Z.-R., Zheng, X.-F., Luo, B.-S., Shen, L.-R., Li, D.-F., Liu, H.-L. & Zhao, W. (2001).Polyhedron,20, 2813–2821.
Oki, A. R., Bommarreddy, P. R., Zhang, H.-M. & Hosmane, N. (1995).Inorg. Chim. Acta,231, 109–114.
Sheldrick, G. M. (1997). SHELXS97 and SHELXL97. University of Go¨ttingen, Germany.
Yan, X.-X., Lu, L.-P. & Zhu, M.-L. (2004).Acta Cryst.C60, m221–m223.
metal-organic papers
m1294
Zhou and Yang [MnCl(Csupporting information
sup-1
Acta Cryst. (2005). E61, m1292–m1294
supporting information
Acta Cryst. (2005). E61, m1292–m1294 [https://doi.org/10.1107/S1600536805017678]
Bis[bis(benzimidazol-2-ylmethyl)amine]-
κ
3N
,
N
′
,
N
′′
;
κ
2N
,
N
′
-chloromanganese(II)
chloride methanol trisolvate
Qing-Hua Zhou and Pin Yang
dichloro[bis(bis(benzimidazol-2-ylmethyl)-amine)] manganese(II) methanol trisolvate
Crystal data
[Mn(C16H15N5)2Cl]Cl·3CH4O
Mr = 776.63 Monoclinic, P21/c
Hall symbol: -P 2ybc
a = 13.161 (3) Å
b = 11.905 (3) Å
c = 25.144 (7) Å
β = 98.794 (4)°
V = 3893.2 (17) Å3
Z = 4
F(000) = 1620
Dx = 1.325 Mg m−3
Mo Kα radiation, λ = 0.71073 Å
Cell parameters from 2271 reflections
θ = 2.3–20.7°
µ = 0.52 mm−1
T = 203 K
Block, colorless 0.40 × 0.30 × 0.10 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
φ and ω scans
Absorption correction: multi-scan (SADABS; Bruker, 1998) Tmin = 0.818, Tmax = 0.950
15680 measured reflections 6860 independent reflections 3596 reflections with I > 2σ(I) Rint = 0.051
θmax = 25.0°, θmin = 1.6°
h = −15→7
k = −14→14
l = −29→29
Refinement
Refinement on F2
Least-squares matrix: full R[F2 > 2σ(F2)] = 0.049
wR(F2) = 0.071
S = 0.93
6860 reflections 490 parameters 4 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H atoms treated by a mixture of independent and constrained refinement
w = 1/[σ2(F
o2) + (0.0102P)2]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max = 0.001
Δρmax = 0.48 e Å−3
supporting information
sup-2
Acta Cryst. (2005). E61, m1292–m1294
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
C1 0.6249 (3) 0.8793 (3) 0.55043 (15) 0.0453 (9)
C2 0.6099 (3) 0.9134 (3) 0.49718 (16) 0.0597 (11)
H2 0.6233 0.9868 0.4876 0.072*
C3 0.5744 (3) 0.8344 (3) 0.45905 (15) 0.0660 (12)
H3 0.5633 0.8543 0.4229 0.079*
C4 0.5547 (3) 0.7242 (3) 0.47399 (16) 0.0585 (11)
H4 0.5302 0.6723 0.4475 0.070*
C5 0.5705 (2) 0.6912 (3) 0.52646 (15) 0.0492 (10)
H5 0.5571 0.6178 0.5359 0.059*
C6 0.6070 (2) 0.7697 (3) 0.56544 (15) 0.0407 (9)
C7 0.6581 (2) 0.8611 (3) 0.63816 (14) 0.0395 (9)
C8 0.6802 (2) 0.8936 (2) 0.69596 (13) 0.0436 (9)
H8A 0.7388 0.9440 0.7015 0.052*
H8B 0.6215 0.9332 0.7059 0.052*
C9 0.6501 (2) 0.7944 (2) 0.77793 (13) 0.0445 (10)
H9A 0.6493 0.8700 0.7923 0.053*
H9B 0.6865 0.7461 0.8055 0.053*
C10 0.5426 (3) 0.7532 (3) 0.76213 (14) 0.0374 (9)
C11 0.3795 (3) 0.7222 (3) 0.76310 (15) 0.0444 (10)
C12 0.2777 (3) 0.7177 (3) 0.77305 (17) 0.0610 (12)
H12 0.2564 0.7557 0.8017 0.073*
C13 0.2113 (3) 0.6540 (3) 0.73801 (19) 0.0654 (13)
H13 0.1431 0.6487 0.7433 0.079*
C14 0.2418 (3) 0.5971 (3) 0.69497 (16) 0.0576 (11)
H14 0.1937 0.5564 0.6718 0.069*
C15 0.3434 (3) 0.6003 (3) 0.68610 (14) 0.0468 (10)
H15 0.3647 0.5614 0.6577 0.056*
C16 0.4120 (3) 0.6639 (3) 0.72153 (14) 0.0378 (9)
C17 0.6966 (3) 0.4826 (2) 0.79777 (14) 0.0356 (9)
C18 0.6077 (2) 0.4871 (2) 0.82103 (14) 0.0433 (10)
H18 0.5507 0.5279 0.8053 0.052*
C19 0.6067 (3) 0.4292 (3) 0.86788 (15) 0.0502 (10)
H19 0.5475 0.4310 0.8838 0.060*
C20 0.6904 (3) 0.3681 (3) 0.89263 (14) 0.0587 (11)
supporting information
sup-3
Acta Cryst. (2005). E61, m1292–m1294
C21 0.7805 (3) 0.3642 (3) 0.87081 (15) 0.0543 (10)
H21 0.8377 0.3246 0.8872 0.065*
C22 0.7809 (3) 0.4226 (2) 0.82315 (14) 0.0383 (9)
C23 0.8139 (3) 0.4950 (3) 0.74777 (14) 0.0372 (9)
C24 0.8708 (3) 0.5171 (3) 0.70236 (15) 0.0628 (12)
H24A 0.9349 0.5549 0.7158 0.075*
H24B 0.8873 0.4462 0.6867 0.075*
C25 0.8115 (3) 0.5409 (3) 0.60666 (14) 0.0570 (11)
H25A 0.7752 0.5929 0.5808 0.068*
H25B 0.7731 0.4710 0.6037 0.068*
C26 0.9152 (3) 0.5187 (4) 0.59121 (15) 0.0532 (11)
C27 1.0561 (3) 0.4407 (4) 0.57943 (16) 0.0643 (12)
C28 1.1382 (4) 0.3681 (4) 0.57664 (17) 0.0949 (16)
H28 1.1361 0.2933 0.5871 0.114*
C29 1.2226 (4) 0.4124 (5) 0.5577 (2) 0.111 (2)
H29 1.2792 0.3665 0.5561 0.133*
C30 1.2260 (4) 0.5224 (5) 0.5410 (2) 0.1027 (19)
H30 1.2842 0.5484 0.5280 0.123*
C31 1.1458 (4) 0.5939 (4) 0.54304 (16) 0.0893 (16)
H31 1.1478 0.6682 0.5319 0.107*
C32 1.0615 (3) 0.5507 (4) 0.56254 (15) 0.0614 (12)
C33 0.7105 (3) 0.2148 (4) 0.55188 (19) 0.1216 (19)
H33A 0.7101 0.2912 0.5639 0.182*
H33B 0.6438 0.1954 0.5331 0.182*
H33C 0.7606 0.2064 0.5281 0.182*
C34 0.9950 (3) 0.1629 (4) 0.6639 (2) 0.137 (2)
H34A 1.0308 0.2026 0.6944 0.205*
H34B 1.0406 0.1502 0.6382 0.205*
H34C 0.9711 0.0922 0.6755 0.205*
C35 0.9077 (4) 0.9133 (4) 0.5394 (2) 0.159 (3)
H35A 0.8654 0.9674 0.5540 0.239*
H35B 0.8755 0.8918 0.5040 0.239*
H35C 0.9738 0.9458 0.5374 0.239*
Cl1 0.07178 (7) 0.32571 (8) 0.80441 (5) 0.0847 (4)
Cl2 0.55819 (7) 0.47079 (7) 0.62765 (4) 0.0553 (3)
H1N 0.7725 (19) 0.791 (2) 0.7422 (11) 0.036 (10)*
H3N 0.679 (2) 1.0056 (12) 0.6049 (11) 0.047 (11)*
H5N 0.464 (2) 0.8301 (19) 0.8147 (9) 0.063 (13)*
H6N 0.844 (2) 0.655 (2) 0.6617 (12) 0.059 (12)*
H8N 0.9177 (12) 0.403 (2) 0.7951 (13) 0.069 (13)*
H10N 0.952 (2) 0.6689 (13) 0.5607 (12) 0.052 (12)*
Mn 0.64222 (4) 0.62451 (4) 0.68032 (2) 0.03878 (15)
N1 0.7022 (2) 0.7940 (2) 0.73029 (12) 0.0384 (8)
N2 0.62919 (19) 0.7602 (2) 0.62102 (11) 0.0383 (7)
N3 0.6569 (2) 0.9362 (2) 0.59732 (13) 0.0475 (8)
N4 0.5163 (2) 0.6842 (2) 0.72148 (11) 0.0365 (7)
N5 0.4643 (3) 0.7780 (2) 0.78885 (13) 0.0465 (8)
supporting information
sup-4
Acta Cryst. (2005). E61, m1292–m1294
N7 0.71986 (19) 0.52869 (19) 0.74986 (11) 0.0364 (7)
N8 0.8551 (2) 0.4332 (2) 0.79024 (13) 0.0424 (8)
N9 0.9631 (2) 0.4215 (2) 0.59725 (12) 0.0599 (9)
N10 0.9697 (3) 0.5982 (3) 0.57086 (13) 0.0613 (10)
O1 0.7341 (3) 0.1478 (3) 0.59372 (14) 0.1097 (12)
H1 0.7924 0.1621 0.6088 0.164*
O2 0.9123 (2) 0.2258 (3) 0.64050 (18) 0.1260 (13)
H2A 0.9316 0.2722 0.6201 0.189*
O3 0.9195 (4) 0.8228 (3) 0.57115 (15) 0.1529 (16)
H3A 0.9244 0.8424 0.6027 0.229*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
C1 0.048 (2) 0.041 (2) 0.048 (3) −0.0013 (19) 0.012 (2) −0.003 (2)
C2 0.077 (3) 0.047 (3) 0.056 (3) −0.003 (2) 0.014 (3) 0.010 (2)
C3 0.085 (3) 0.067 (3) 0.047 (3) 0.003 (2) 0.012 (2) 0.002 (3)
C4 0.071 (3) 0.052 (3) 0.050 (3) 0.001 (2) 0.005 (2) −0.010 (2)
C5 0.053 (3) 0.042 (2) 0.052 (3) 0.0030 (19) 0.008 (2) −0.002 (2)
C6 0.039 (2) 0.037 (2) 0.048 (3) 0.0006 (18) 0.010 (2) −0.002 (2)
C7 0.033 (2) 0.042 (2) 0.044 (3) −0.0021 (18) 0.0086 (19) −0.002 (2)
C8 0.045 (2) 0.035 (2) 0.052 (3) −0.0030 (17) 0.011 (2) −0.003 (2)
C9 0.053 (3) 0.035 (2) 0.047 (3) −0.0011 (18) 0.010 (2) −0.0060 (18)
C10 0.040 (3) 0.030 (2) 0.044 (3) 0.0061 (18) 0.012 (2) 0.0034 (18)
C11 0.041 (3) 0.037 (2) 0.057 (3) 0.011 (2) 0.015 (2) 0.011 (2)
C12 0.056 (3) 0.052 (3) 0.081 (4) 0.018 (2) 0.031 (3) 0.023 (2)
C13 0.041 (3) 0.064 (3) 0.096 (4) 0.015 (2) 0.027 (3) 0.036 (3)
C14 0.038 (3) 0.055 (3) 0.076 (3) −0.003 (2) −0.002 (2) 0.026 (2)
C15 0.040 (2) 0.045 (2) 0.054 (3) 0.0051 (19) 0.005 (2) 0.014 (2)
C16 0.034 (2) 0.032 (2) 0.047 (3) 0.0031 (18) 0.007 (2) 0.0098 (18)
C17 0.030 (2) 0.031 (2) 0.047 (2) −0.0014 (17) 0.010 (2) −0.0067 (18)
C18 0.038 (2) 0.041 (2) 0.052 (3) 0.0017 (18) 0.010 (2) 0.000 (2)
C19 0.053 (3) 0.050 (2) 0.052 (3) 0.001 (2) 0.020 (2) −0.001 (2)
C20 0.065 (3) 0.060 (3) 0.052 (3) 0.007 (2) 0.013 (2) 0.015 (2)
C21 0.047 (3) 0.055 (3) 0.059 (3) 0.012 (2) 0.003 (2) 0.004 (2)
C22 0.040 (2) 0.031 (2) 0.044 (3) −0.0017 (18) 0.008 (2) −0.0034 (18)
C23 0.035 (2) 0.036 (2) 0.041 (2) 0.0014 (18) 0.009 (2) −0.0009 (18)
C24 0.041 (3) 0.084 (3) 0.066 (3) 0.014 (2) 0.016 (2) 0.010 (2)
C25 0.048 (3) 0.060 (3) 0.065 (3) 0.009 (2) 0.014 (2) −0.008 (2)
C26 0.046 (3) 0.065 (3) 0.052 (3) 0.002 (2) 0.018 (2) −0.009 (2)
C27 0.051 (3) 0.080 (3) 0.067 (3) 0.012 (3) 0.026 (2) −0.008 (3)
C28 0.075 (4) 0.116 (4) 0.102 (4) 0.030 (3) 0.039 (3) −0.008 (3)
C29 0.064 (4) 0.183 (6) 0.095 (4) 0.031 (4) 0.040 (3) −0.007 (4)
C30 0.073 (4) 0.160 (6) 0.085 (4) −0.022 (4) 0.043 (3) −0.020 (4)
C31 0.087 (4) 0.112 (4) 0.075 (4) −0.022 (3) 0.033 (3) −0.029 (3)
C32 0.051 (3) 0.084 (3) 0.055 (3) −0.010 (3) 0.027 (2) −0.017 (3)
C33 0.133 (5) 0.097 (4) 0.125 (5) −0.024 (3) −0.010 (4) 0.019 (4)
supporting information
sup-5
Acta Cryst. (2005). E61, m1292–m1294
C35 0.165 (6) 0.123 (5) 0.203 (7) 0.011 (4) 0.073 (5) 0.062 (5)
Cl1 0.0486 (7) 0.0804 (8) 0.1281 (11) 0.0251 (6) 0.0233 (7) 0.0153 (7)
Cl2 0.0677 (7) 0.0433 (5) 0.0553 (7) −0.0179 (5) 0.0110 (6) −0.0062 (5)
Mn 0.0354 (3) 0.0341 (3) 0.0476 (4) −0.0010 (3) 0.0090 (3) −0.0029 (3)
N1 0.029 (2) 0.0389 (18) 0.045 (2) −0.0045 (15) −0.0011 (17) 0.0001 (15)
N2 0.0393 (18) 0.0322 (17) 0.043 (2) −0.0033 (14) 0.0031 (15) −0.0074 (15)
N3 0.058 (2) 0.030 (2) 0.055 (2) −0.0104 (17) 0.0117 (18) −0.0024 (19)
N4 0.0288 (18) 0.0346 (17) 0.046 (2) −0.0015 (13) 0.0043 (15) 0.0027 (15)
N5 0.053 (2) 0.038 (2) 0.053 (2) 0.0070 (17) 0.020 (2) −0.0006 (18)
N6 0.045 (2) 0.0368 (19) 0.048 (2) −0.0028 (16) 0.0147 (17) −0.0011 (17)
N7 0.0276 (18) 0.0359 (16) 0.048 (2) 0.0037 (13) 0.0118 (15) −0.0020 (15)
N8 0.031 (2) 0.0453 (19) 0.051 (2) 0.0102 (16) 0.0065 (18) −0.0012 (16)
N9 0.048 (2) 0.063 (2) 0.073 (3) 0.0107 (18) 0.025 (2) −0.0060 (19)
N10 0.071 (3) 0.060 (3) 0.059 (2) 0.001 (2) 0.029 (2) −0.005 (2)
O1 0.141 (3) 0.074 (2) 0.102 (3) −0.048 (2) −0.019 (2) 0.032 (2)
O2 0.099 (3) 0.081 (3) 0.188 (4) −0.0103 (19) −0.011 (3) 0.035 (2)
O3 0.258 (4) 0.073 (2) 0.132 (4) 0.022 (3) 0.045 (4) 0.000 (2)
Geometric parameters (Å, º)
C1—N3 1.369 (4) C23—C24 1.482 (4)
C1—C2 1.384 (4) C24—N6 1.467 (4)
C1—C6 1.387 (4) C24—H24A 0.9700
C2—C3 1.374 (4) C24—H24B 0.9700
C2—H2 0.9300 C25—N6 1.466 (4)
C3—C4 1.399 (4) C25—C26 1.499 (4)
C3—H3 0.9300 C25—H25A 0.9700
C4—C5 1.362 (4) C25—H25B 0.9700
C4—H4 0.9300 C26—N9 1.316 (4)
C5—C6 1.386 (4) C26—N10 1.336 (4)
C5—H5 0.9300 C27—C32 1.381 (4)
C6—N2 1.388 (4) C27—N9 1.385 (4)
C7—N2 1.313 (3) C27—C28 1.394 (5)
C7—N3 1.359 (4) C28—C29 1.380 (5)
C7—C8 1.489 (4) C28—H28 0.9300
C8—N1 1.469 (3) C29—C30 1.377 (5)
C8—H8A 0.9700 C29—H29 0.9300
C8—H8B 0.9700 C30—C31 1.364 (5)
C9—N1 1.469 (4) C30—H30 0.9300
C9—C10 1.492 (4) C31—C32 1.379 (4)
C9—H9A 0.9700 C31—H31 0.9300
C9—H9B 0.9700 C32—N10 1.379 (4)
C10—N4 1.316 (4) C33—O1 1.318 (4)
C10—N5 1.346 (4) C33—H33A 0.9600
C11—N5 1.373 (4) C33—H33B 0.9600
C11—C16 1.376 (4) C33—H33C 0.9600
C11—C12 1.401 (4) C34—O2 1.377 (4)
supporting information
sup-6
Acta Cryst. (2005). E61, m1292–m1294
C12—H12 0.9300 C34—H34B 0.9600
C13—C14 1.387 (4) C34—H34C 0.9600
C13—H13 0.9300 C35—O3 1.336 (4)
C14—C15 1.389 (4) C35—H35A 0.9600
C14—H14 0.9300 C35—H35B 0.9600
C15—C16 1.392 (4) C35—H35C 0.9600
C15—H15 0.9300 Mn—Cl2 2.425 (1)
C16—N4 1.394 (3) Mn—N1 2.444 (3)
C17—C18 1.387 (4) Mn—N2 2.187 (3)
C17—C22 1.390 (4) Mn—N4 2.202 (2)
C17—N7 1.400 (4) Mn—N6 2.389 (3)
C18—C19 1.366 (4) Mn—N7 2.204 (3)
C18—H18 0.9300 N1—H1N 0.93 (2)
C19—C20 1.386 (4) N3—H3N 0.888 (10)
C19—H19 0.9300 N5—H5N 0.898 (10)
C20—C21 1.381 (4) N6—H6N 0.92 (3)
C20—H20 0.9300 N8—H8N 0.890 (10)
C21—C22 1.386 (4) N10—H10N 0.899 (10)
C21—H21 0.9300 O1—H1 0.8200
C22—N8 1.379 (4) O2—H2A 0.8200
C23—N7 1.309 (3) O3—H3A 0.8200
C23—N8 1.341 (4)
N3—C1—C2 131.8 (4) N10—C26—C25 122.5 (4)
N3—C1—C6 105.9 (3) C32—C27—N9 110.2 (4)
C2—C1—C6 122.3 (4) C32—C27—C28 119.7 (4)
C3—C2—C1 117.2 (3) N9—C27—C28 130.1 (5)
C3—C2—H2 121.4 C29—C28—C27 116.9 (5)
C1—C2—H2 121.4 C29—C28—H28 121.6
C2—C3—C4 120.8 (4) C27—C28—H28 121.6
C2—C3—H3 119.6 C30—C29—C28 122.3 (5)
C4—C3—H3 119.6 C30—C29—H29 118.9
C5—C4—C3 121.6 (4) C28—C29—H29 118.9
C5—C4—H4 119.2 C31—C30—C29 121.4 (5)
C3—C4—H4 119.2 C31—C30—H30 119.3
C4—C5—C6 118.4 (3) C29—C30—H30 119.3
C4—C5—H5 120.8 C30—C31—C32 116.8 (5)
C6—C5—H5 120.8 C30—C31—H31 121.6
C5—C6—C1 119.8 (4) C32—C31—H31 121.6
C5—C6—N2 130.8 (3) C31—C32—N10 132.2 (5)
C1—C6—N2 109.4 (3) C31—C32—C27 123.0 (4)
N2—C7—N3 112.6 (3) N10—C32—C27 104.8 (4)
N2—C7—C8 124.2 (3) O1—C33—H33A 109.5
N3—C7—C8 123.0 (3) O1—C33—H33B 109.5
N1—C8—C7 110.8 (3) H33A—C33—H33B 109.5
N1—C8—H8A 109.5 O1—C33—H33C 109.5
C7—C8—H8A 109.5 H33A—C33—H33C 109.5
supporting information
sup-7
Acta Cryst. (2005). E61, m1292–m1294
C7—C8—H8B 109.5 O2—C34—H34A 109.5
H8A—C8—H8B 108.1 O2—C34—H34B 109.5
N1—C9—C10 108.9 (3) H34A—C34—H34B 109.5
N1—C9—H9A 109.9 O2—C34—H34C 109.5
C10—C9—H9A 109.9 H34A—C34—H34C 109.5
N1—C9—H9B 109.9 H34B—C34—H34C 109.5
C10—C9—H9B 109.9 O3—C35—H35A 109.5
H9A—C9—H9B 108.3 O3—C35—H35B 109.5
N4—C10—N5 113.2 (3) H35A—C35—H35B 109.5
N4—C10—C9 122.6 (3) O3—C35—H35C 109.5
N5—C10—C9 124.2 (3) H35A—C35—H35C 109.5
N5—C11—C16 106.1 (3) H35B—C35—H35C 109.5
N5—C11—C12 131.7 (4) N2—Mn—N4 95.50 (9)
C16—C11—C12 122.2 (4) N2—Mn—N7 154.4 (1)
C13—C12—C11 116.1 (4) N4—Mn—N7 94.9 (1)
C13—C12—H12 122.0 N2—Mn—N6 88.7 (1)
C11—C12—H12 122.0 N4—Mn—N6 161.39 (11)
C12—C13—C14 122.6 (4) N7—Mn—N6 74.5 (1)
C12—C13—H13 118.7 N2—Mn—Cl2 101.73 (8)
C14—C13—H13 118.7 N4—Mn—Cl2 100.79 (8)
C13—C14—C15 120.9 (4) N7—Mn—Cl2 99.22 (7)
C13—C14—H14 119.6 N6—Mn—Cl2 96.06 (8)
C15—C14—H14 119.6 N2—Mn—N1 74.1 (1)
C14—C15—C16 117.2 (3) N4—Mn—N1 72.5 (1)
C14—C15—H15 121.4 N7—Mn—N1 86.9 (1)
C16—C15—H15 121.4 N6—Mn—N1 91.4 (1)
C11—C16—C15 120.9 (4) Cl2—Mn—N1 171.39 (8)
C11—C16—N4 109.5 (3) C9—N1—C8 113.7 (3)
C15—C16—N4 129.5 (3) C9—N1—Mn 105.52 (18)
C18—C17—C22 119.5 (3) C8—N1—Mn 110.2 (2)
C18—C17—N7 131.1 (3) C9—N1—H1N 107.7 (17)
C22—C17—N7 109.4 (3) C8—N1—H1N 108.3 (16)
C19—C18—C17 117.6 (3) Mn—N1—H1N 111.5 (16)
C19—C18—H18 121.2 C7—N2—C6 105.2 (3)
C17—C18—H18 121.2 C7—N2—Mn 117.6 (2)
C18—C19—C20 122.7 (3) C6—N2—Mn 136.9 (2)
C18—C19—H19 118.6 C7—N3—C1 107.0 (3)
C20—C19—H19 118.6 C7—N3—H3N 119 (2)
C21—C20—C19 120.7 (3) C1—N3—H3N 134 (2)
C21—C20—H20 119.6 C10—N4—C16 104.5 (3)
C19—C20—H20 119.6 C10—N4—Mn 116.1 (2)
C20—C21—C22 116.3 (3) C16—N4—Mn 139.1 (2)
C20—C21—H21 121.8 C10—N5—C11 106.7 (3)
C22—C21—H21 121.8 C10—N5—H5N 127 (2)
N8—C22—C21 131.4 (3) C11—N5—H5N 126 (2)
N8—C22—C17 105.5 (3) C25—N6—C24 111.9 (3)
C21—C22—C17 123.1 (3) C25—N6—Mn 113.5 (2)
supporting information
sup-8
Acta Cryst. (2005). E61, m1292–m1294
N7—C23—C24 124.7 (3) C25—N6—H6N 106.5 (19)
N8—C23—C24 121.4 (3) C24—N6—H6N 106 (2)
N6—C24—C23 111.7 (3) Mn—N6—H6N 106.1 (17)
N6—C24—H24A 109.3 C23—N7—C17 104.3 (3)
C23—C24—H24A 109.3 C23—N7—Mn 116.9 (2)
N6—C24—H24B 109.3 C17—N7—Mn 138.6 (2)
C23—C24—H24B 109.3 C23—N8—C22 106.9 (3)
H24A—C24—H24B 107.9 C23—N8—H8N 126 (2)
N6—C25—C26 116.1 (3) C22—N8—H8N 127 (2)
N6—C25—H25A 108.3 C26—N9—C27 104.4 (3)
C26—C25—H25A 108.3 C26—N10—C32 107.5 (4)
N6—C25—H25B 108.3 C26—N10—H10N 130.0 (19)
C26—C25—H25B 108.3 C32—N10—H10N 122.2 (19)
H25A—C25—H25B 107.4 C33—O1—H1 109.5
N9—C26—N10 113.1 (3) C34—O2—H2A 109.5
N9—C26—C25 124.4 (4) C35—O3—H3A 109.5
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
O1—H1···O2 0.82 1.82 2.627 (4) 167
O2—H2A···N9 0.82 1.93 2.698 (4) 155
O3—H3A···Cl1i 0.82 2.34 3.114 (4) 158
N1—H1N···Cl1i 0.93 (2) 2.54 (2) 3.247 (3) 133 (2)
N3—H3N···O1ii 0.89 (1) 1.88 (1) 2.724 (4) 158 (3)
N5—H5N···Cl2i 0.90 (1) 2.26 (1) 3.155 (3) 171 (3)
N6—H6N···Cl1i 0.92 (3) 2.41 (3) 3.292 (3) 161 (3)
N8—H8N···Cl1iii 0.89 (1) 2.21 (1) 3.096 (3) 178 (3)
N10—H10N···O3 0.90 (1) 1.91 (2) 2.754 (4) 156 (3)
C4—H4···Cl2iv 0.93 2.67 3.603 (4) 174