(2
R
)-2-Cinnamoylamino-
N
-[5-(4-
methoxyphenyl)-1,3,4-thiadiazol-2-yl]-propanamide
Shao-Hua Li,a,bGang Li,bHui-Ming Huang,bGuo-Gang Tub* and Cheng-Mei Liua‡
aState Key Laboratory of Food Science and Technology, Nanchang University, 330047 Nanchang, JiangXi, People’s Republic of China, andbDepartment of Pharmacy, NanChang University Medical College, 330006 Nanchang, JiangXi, People’s Republic of China
Correspondence e-mail: tugg199@yahoo.com
Received 29 July 2008; accepted 30 August 2008
Key indicators: single-crystal X-ray study;T= 296 K; mean(C–C) = 0.007 A˚;
Rfactor = 0.039;wRfactor = 0.098; data-to-parameter ratio = 6.7.
The asymmetric unit of the title compound, C21H20N4O3S, contains two independent molecules. The dihedral angles between the two benzene rings are 47.6 (1) and 30.2 (1), the
corresponding values between the p-methoxybenzene and thiadiazol rings are 12.3 (1) and 24.7 (1), respectively, for the
two molecules. The conformations of the N—H and C O bonds areantiwith respect to each other. The enone groups show atransconfiguration. The crystal structure is stabilized by N—H O and N—H N interactions. The absolute structure could not be determined from the X-ray data but the absolute configuration has been assigned by reference to an unchanging chiral centre in the synthetic procedure.
Related literature
For 1,3,4-thiadiazole scaffold compounds and their biological activity, see: Tuet al.(2008). For the synthesis, see: Foroumadi
et al. (1999); Levy & Palmer (1942); Song et al. (1992). For related structures, see: Fun et al.(2008); Gowda et al.(2008) Thiruvalluvaret al.(2008).
Experimental
Crystal data
C21H20N4O3S Mr= 408.48
Triclinic,P1
a= 9.082 (3) A˚
b= 9.849 (3) A˚
c= 13.644 (4) A˚ = 79.587 (4)
= 83.253 (4)
= 65.458 (4)
V= 1090.8 (5) A˚3 Z= 2
MoKradiation = 0.18 mm1 T= 296 (2) K 0.350.240.06 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: none 6219 measured reflections
3553 independent reflections 2735 reflections withI> 2(I)
Rint= 0.022
Refinement
R[F2> 2(F2)] = 0.038 wR(F2) = 0.097
S= 0.94 3553 reflections 528 parameters
3 restraints
H-atom parameters constrained
max= 0.20 e A˚
3
min=0.17 e A˚
[image:1.610.313.564.309.365.2]3
Table 1
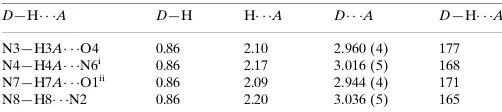
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
N3—H3A O4 0.86 2.10 2.960 (4) 177 N4—H4A N6i
0.86 2.17 3.016 (5) 168 N7—H7A O1ii
0.86 2.09 2.944 (4) 171 N8—H8 N2 0.86 2.20 3.036 (5) 165
Symmetry codes: (i)x;y1;z; (ii)x;yþ1;z.
Data collection:APEX2(Bruker, 2004); cell refinement:APEX2 andSAINT(Bruker, 2004); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 2008); program(s) used to refine structure:SHELXL97(Sheldrick, 2008); molecular graphics: APEX2; software used to prepare material for publication:APEX2 andpublCIF(Westrip, 2008).
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IM2079).
References
Bruker (2004).APEX2andSAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
Foroumadi, A., Daneshtalab, M. & Shafiee, A. (1999).Arzneim. Forsch.49, 1035–1038.
Fun, H.-K., Chantrapromma, S., Patil, P. S., Karthikeyan, M. S. & Dharmaprakash, S. M. (2008).Acta Cryst.E64, o956–o957.
Gowda, B. T., Tokarcˇı´k, M., Kozˇı´sˇek, J., Sowmya, B. P. & Fuess, H. (2008).Acta Cryst.E64, o950.
Levy, M. & Palmer, A. H. (1942).J. Biol. Chem.146, 493–495. Sheldrick, G. M. (2008).Acta Cryst.A64, 112–122.
Song, K. S., Ishikawa, Y., Kobayashi, S., Sankawa, U. & Ebizuka, Y. (1992).
Phytochemistry,31, 823–826.
Thiruvalluvar, A., Subramanyam, M., Butcher, R. J., Karabasanagouda, T. & Adhikari, A. V. (2008).Acta Cryst.E64, o1263.
Tu, G. G., Li, S. H., Huang, H. M., Li, G., Xiong, F., Mai, X., Zhu, H. W., Kuang, B. H. & Xu, W. F. (2008).Bioorg. Med. Chem.16, 6663–6668.
Westrip, S. P. (2008).publCIF. In preparation. Acta Crystallographica Section E
Structure Reports
Online
supporting information
Acta Cryst. (2008). E64, o1887 [doi:10.1107/S1600536808027803]
(2
R
)-2-Cinnamoylamino-
N
-[5-(4-methoxyphenyl)-1,3,4-thiadiazol-2-yl]propanamide
Shao-Hua Li, Gang Li, Hui-Ming Huang, Guo-Gang Tu and Cheng-Mei Liu
S1. Comment
In our previous work, 1,3,4-thiadiazole scaffold compounds and their biological activity have been studied (Tu et al., 2008). In view of the importance of these organic materials, the title compound (Fig. 1) was synthesized (Foroumadi et al., 1999; Levy & Palmer 1942; Song et al., 1992) and its crystal structure is reported here. The asymmetric unit of the title compound, C21H20N4O3S, contains two independent molecules. The dihedral angles between the p-methoxybenzene
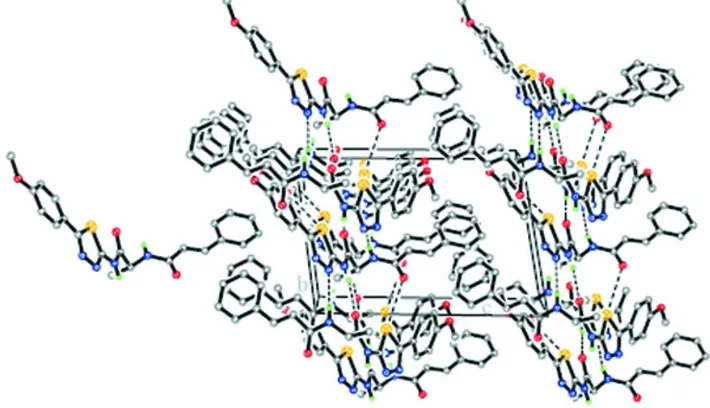
and thiadiazol rings is 12.2 (1) ° and 24.7 (1)°, the corresponding values between the two benzene rings are measured to 47.6 (1)° and 30.2 (1)°, respectively, for the two molecules. The conformations of the N—H and C=O bonds are anti with respect to each other. The enone groups are trans configurated. Bond lengths and angles are in normal ranges and comparable to those in related structures (Gowda et al., 2008; Fun et al., 2008; Thiruvalluvar et al., 2008). In the crystal structure, molecules are linked through intermolecular N—H···O hydrogen bonds forming a three-dimensional network (Table 1, Figure 2).
S2. Experimental
N,Dicyclohexylcarbodiimide (5.7 mmol) was added to a cooled solution of cinnamoyl-D-alanine (5.6 mmol) and N-hydroxysuccinimide (5.6 mmol) in freshly distilled dioxane (30 ml). The reaction mixture was stirred overnight at room temperature. The insoluble material was filtered off and washed with cold dioxane. 2-Amino-5-(4-methoxyphenyl)-1,3,4-thiadiazole (5.5 mmol) was added to the filtrate and the reaction mixture was stirred for 48 hr at room temperature. The solvent was removed under reduced pressure. The residual was dissolved in EtOAc and the insoluble material was filtered off. The filtrate was washed successively with saturated Na2CO3 solution (20 ml, x 3), water (20 ml, x 1), 0.1 M
HCl (20 ml, x 3) and water (20 ml, x 1). The organic layer was evaporated in vacuo and the residue was recrystallized from methanol (30 ml), yield: 35.2%. Colorless block-shaped single crystals of the title compound suitable for X-ray diffraction analysis precipitated after several days.
S3. Refinement
H atoms were positioned geometrically and refined using a riding model using SHELXL97 default values (Uĩso(H) = 1.2
Ueq(C) for CH and CH2 groups and Uĩso(H) = 1.5 Ueq(C) for CH3). Refinement with all data (Friedel opposites not merged)
Figure 1
Molecular structure of the two independent molecules of the title compound. Displacement ellipsoids are drawn at the 50% probability level.
Figure 2
The crystal packing of title compound, viewed along the a axis with hydrogen bonds drawn as dashed lines.
(2R)-2-Cinnamoylamino-N-[5-(4-methoxyphenyl) -1,3,4-thiadiazol-2-yl]propanamide
Crystal data C21H20N4O3S
Mr = 408.48
Triclinic, P1 Hall symbol: P 1 a = 9.082 (3) Å b = 9.849 (3) Å c = 13.644 (4) Å α = 79.587 (4)° β = 83.253 (4)° γ = 65.458 (4)° V = 1090.8 (5) Å3
Z = 2 F(000) = 428 Dx = 1.244 Mg m−3
Melting point: 450 K
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 2616 reflections θ = 2.3–25.0°
µ = 0.18 mm−1
[image:3.610.126.481.307.511.2]Data collection
Bruker APEXII CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
phi and ω scans
6219 measured reflections 3553 independent reflections
2735 reflections with I > 2σ(I) Rint = 0.022
θmax = 25.0°, θmin = 2.3°
h = −10→10 k = −11→11 l = −16→16
Refinement Refinement on F2
Least-squares matrix: full R[F2 > 2σ(F2)] = 0.038
wR(F2) = 0.097
S = 0.94 3553 reflections 528 parameters 3 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained w = 1/[σ2(F
o2) + (0.0648P)2]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.20 e Å−3
Δρmin = −0.18 e Å−3
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. # SQUEEZE RESULTS (APPEND TO CIF) loop_ _platon_squeeze_void_nr _platon_squeeze_void_average_x _platon_squeeze_void_average_y _platon_squeeze_void_average_z _platon_squeeze_void_volume
_platon_squeeze_void_count_electrons 1 0.151 0.591 0.943 19.4 0.7 2 0.269 -0.200 0.408 64.4 11.5
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
C35 −0.2660 (5) 0.6200 (5) 0.4430 (3) 0.0636 (11) H35 −0.3026 0.5591 0.4171 0.076* C36 −0.3019 (5) 0.6389 (5) 0.5363 (3) 0.0638 (11) H36 −0.2649 0.7022 0.5589 0.077* C37 −0.3926 (5) 0.5732 (5) 0.6091 (3) 0.0626 (11) C38 −0.4185 (7) 0.6053 (7) 0.7051 (4) 0.0899 (16) H38 −0.3807 0.6716 0.7228 0.108* C39 −0.5020 (8) 0.5383 (9) 0.7766 (4) 0.105 (2) H39 −0.5189 0.5604 0.8415 0.126* C40 −0.5591 (7) 0.4401 (7) 0.7516 (5) 0.0953 (18) H40 −0.6158 0.3970 0.7988 0.114* C41 −0.5316 (8) 0.4077 (7) 0.6580 (5) 0.0948 (17) H41 −0.5690 0.3408 0.6408 0.114* C42 −0.4493 (6) 0.4714 (6) 0.5874 (4) 0.0807 (14) H42 −0.4310 0.4459 0.5233 0.097* N1 −0.3581 (4) 0.3920 (4) 0.3237 (3) 0.0643 (9) N2 −0.2200 (4) 0.3929 (4) 0.2696 (3) 0.0641 (9) N3 0.0283 (4) 0.2369 (4) 0.2044 (2) 0.0555 (8) H3A 0.0578 0.3109 0.1960 0.067* N4 0.3805 (4) 0.0138 (4) 0.0618 (2) 0.0556 (8) H4A 0.3890 −0.0778 0.0710 0.067* N5 0.6130 (4) 0.5942 (4) 0.0479 (3) 0.0669 (10) N6 0.4668 (4) 0.6790 (4) 0.0918 (3) 0.0668 (10) N7 0.2200 (4) 0.6771 (4) 0.1557 (3) 0.0567 (8) H7A 0.1911 0.7681 0.1667 0.068* N8 −0.1393 (4) 0.6583 (4) 0.2839 (3) 0.0603 (9) H8 −0.1699 0.5930 0.2693 0.072* O1 0.1002 (3) −0.0005 (3) 0.1732 (2) 0.0627 (8) O2 0.4184 (3) 0.2007 (3) −0.0448 (2) 0.0634 (7) O4 0.1420 (3) 0.4837 (3) 0.1726 (2) 0.0652 (8) O5 −0.1232 (5) 0.7795 (4) 0.4039 (2) 0.0849 (10) O3 −0.8515 (5) 0.1357 (5) 0.5771 (4) 0.1156 (15) O6 1.1766 (4) 0.0707 (4) −0.1543 (3) 0.0913 (11) S1 −0.18753 (12) 0.11853 (11) 0.29467 (8) 0.0607 (3) S2 0.45630 (12) 0.42627 (11) 0.08513 (8) 0.0581 (3)
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
S2 0.0546 (6) 0.0436 (5) 0.0817 (7) −0.0264 (5) 0.0176 (5) −0.0201 (5)
Geometric parameters (Å, º)
C19—H19 0.9300 C42—H42 0.9300 C20—C21 1.391 (7) N1—N2 1.381 (5) C20—H20 0.9300 N3—H3A 0.8600 C21—H21 0.9300 N4—H4A 0.8600 C22—O6 1.430 (8) N5—N6 1.376 (5) C22—H22A 0.9600 N7—H7A 0.8600 C22—H22B 0.9600 N8—H8 0.8600
C14—C15—H15 116.3 C38—C39—H39 119.7 C16—C15—H15 116.3 C41—C40—C39 119.0 (5) C17—C16—C21 117.2 (4) C41—C40—H40 120.5 C17—C16—C15 123.3 (4) C39—C40—H40 120.5 C21—C16—C15 119.4 (4) C40—C41—C42 121.1 (6) C18—C17—C16 120.2 (5) C40—C41—H41 119.4 C18—C17—H17 119.9 C42—C41—H41 119.4 C16—C17—H17 119.9 C41—C42—C37 121.7 (5) C19—C18—C17 122.5 (6) C41—C42—H42 119.2 C19—C18—H18 118.7 C37—C42—H42 119.2 C17—C18—H18 118.7 C8—N1—N2 112.0 (3) C18—C19—C20 118.8 (5) C9—N2—N1 112.1 (3) C18—C19—H19 120.6 C9—N3—C10 124.9 (3) C20—C19—H19 120.6 C9—N3—H3A 117.6 C19—C20—C21 120.6 (5) C10—N3—H3A 117.6 C19—C20—H20 119.7 C13—N4—C11 120.7 (3) C21—C20—H20 119.7 C13—N4—H4A 119.7 C16—C21—C20 120.5 (5) C11—N4—H4A 119.7 C16—C21—H21 119.7 C29—N5—N6 112.9 (3) C20—C21—H21 119.7 C30—N6—N5 112.2 (3) O6—C23—C28 125.5 (5) C31—N7—C30 126.0 (3) O6—C23—C24 114.7 (4) C31—N7—H7A 117.0 C28—C23—C24 119.7 (4) C30—N7—H7A 117.0 C25—C24—C23 119.5 (4) C34—N8—C32 122.6 (3) C25—C24—H24 120.2 C34—N8—H8 118.7 C23—C24—H24 120.2 C32—N8—H8 118.7 C24—C25—C26 121.9 (4) C2—O3—C1 117.5 (5) C24—C25—H25 119.0 C23—O6—C22 117.9 (4) C26—C25—H25 119.0 C9—S1—C8 86.3 (2) C27—C26—C25 117.5 (4) C30—S2—C29 86.49 (19)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A N3—H3A···O4 0.86 2.10 2.960 (4) 177 N4—H4A···N6i 0.86 2.17 3.016 (5) 168
N7—H7A···O1ii 0.86 2.09 2.944 (4) 171
N8—H8···N2 0.86 2.20 3.036 (5) 165