SPECIALIZED TRANSDUCING PHAGES DERIVED FROM SALMONELLA PHAGE P22
INGRID HOPPE A N D JOHN ROTH
Department of Molecular Biology, University of California, Berkeley, California 94720
Manuscript received September 4, 1973
ABSTRACT
Salmonella phage P22 has been used in the construction of three sorts of specialized transducing phage: P22 proAB, P22 proABlac and P22 argF. The bacterial genes carried are derived from E. coli K12. Since E. coli and Sal- monella chromosomes recombine very poorly, E. coli genes cannot be trans- duced into Salmonella recipients by P22‘s generalized transduction mechanism. Therefore, stable inheritance of E. coli material provides a means of detecting specialized transduction. Formation of these phages was possible because the
P22 prophage recognizes an attachment site in the E. coli F prolac episome. Salmonella strains carrying the F’ prolac episome can be lysogenized by P22 SO as to leave the prophage inserted into the E. coli material of the F’ factor. Improper prophage excision can then lead to formation of P22 specialized phages carrying E. coli genetic material.
ALMONELLA phage P22 has been widely used in genetic analysis as a generalized transducing phage. The process of generalized transduction (reviewed by OZEKI and IKEDA 1968) involves formation of transducing particles during lytic phage growth. These particles appear to arise when bacterial genes, selected more or less at random, are encapsulated in a phage head. Upon subse- quent infection of a strain by such a particle the bacterial genes are injected and can recombine with the host chromosome. This sort of transduction requires homology between the transduced material and the recipient chromosome; the host recombination system ( m c ) is also required. The recombinants arising by this transduction mode are stable.
Specialized transduction as performed by coliphages A and $30 is a basically different process (reviewed by CAMPBELL 1971). Bacterial genes are incorporated into a phage genome as a consequence of improper prophage excision. Therefore, lysogeny and prophage induction are generally prerequisites for transducing phage formation, Only bacterial genes located near the prophage attachment site are subject to specialized transduction. Since the transduced genes are part of the phage genome, they can be inserted into the recipient chromosome by the phage integration system; this does not require the host rec system. Like a prophage, the transducing phage genome can be lost from the chromosome. This makes the transductant clones unstable in that the inherited genes are subject
to loss.
634 I. HOPPE A N D J. ROTH
Although P22 is best known as a generalized transducing phage, there is reason to expect that phage P22 might also be capable of specialized transduction. This expectation is based on the similarities between P22 and phage h. Both
phages form UV-inducible lysogens in which the prophage inserts at a particular point in the host chromosome. Phage h inserts between the gal and bio operons of E . coli; P22 inserts between the p r o A and proC loci of Salmonella. One might expect that P22, like h, could occasionally make mistakes during prophage excision to form specialized transducing phages.
Several reports indicate that P22 can, in fact, perform specialized transduction
of the p r o A and proB genes. SMITH-KEARY (1966) reported unstable (Pro+) transductants which give rise, when UV-induced, to high-frequency pro trans- ducing lysates. JENNIFER WING (1968) reported that UV-induction of a P22 lysogen gives rise to a lysate which can transduce the p r o A and proB genes into r e d - recipients. Lytically grown P22 does not perform such transduction. These findings are consistent with formation of P22 specialized transducing phage by abnormal excision of the prophage. This specialized phage could then lysogenize at the P22 attachment site and form unstable merodiploids. Such transduction would not be dependent on the rec system of recombination. This course of events would be basically the same as specialized transduction of gal and bio by coli- phage h.
Several recent publications suggest that P22 specialized transducing phage may also be formed by novel means. ADRIENNE JESSOP (1972) has reported formation of specialized phages during lytic growth of P22; these phages are detected in transductional crosses in which the recipient lacks the normal pro- phage attachment site. Thus normal prerequisites of specialized phage formation were absent, yet high frequency transducing lysates could be prepared which seemed to contain specialized transducing phage. A P22 specialized transducing phage for tetracycline resistance ( tetR) has been isolated (DUBNAU and STOCKER
1964;
WATANABE
et al. 1972). The nature of this phage has been investigated i n considerable detail (CHEN et al. 1972). This phage was selected during a trans- duction cross in which the donor strain carried an R-factor with genes for tetra- cycline resistance; the recipient had no R-factor and thus no DNA sequence homologous to tetR which could permit normal recombination of the tetR genes. Selection was made for tetracycline resistance. One recombinant type is a P22 lysogen in which tetracycline resistance is associated with the prophage. ThetetR determinant has been inserted into the prophage map with no loss of P22 material. This is very different from the addition of bacterial genes to the end of the prophage map (and the concomitant loss of phage genes) which is character-
P22 SPECIALIZED TRANSDUCTION
635
Analysis of P22 specialized transduction is difficult in that one must contend with a high background of generalized transduction that accompanies every cross. In the case of the tetR transducing phage, lack of homology between tetR and the recipient chromosome circumvents the problem of generalized trans- duction. Alternatively, one could use recombination-deficient recipients, but this might interfere with some sorts of defective phage particles since some recombi- nation is essential for circularization of the P22 genome (BOTSTEIN and MATZ 1970).
We have circumvented generalized recombination by studying the transduc- tion of E. coli genetic material. The DNA sequence homology between E. coli
and Salmonella is sufficiently low that recombination occurs very rarely. Thus E. coli material is essentially not transducible into Salmonella recipients by generalized transduction. Thus one can search for rare specialized phages which are capable of transducing the E. coli material. The specialized transducing phages we have selected in this way are able to transduce E . coli genes very efficiently from one Salmonella strain to another. They are able to do this since prophage integration, not recombination, is responsible for insertion of the trans- duced material into the recipient chromosome. The formation of P22 specialized transducing phages carrying E. coli material was possible because P22 recognizes a prophage attachment site in the chromosome of E. coli and thus will integrate adjacent to E. coli genetic material. In this paper we present evidence for this attachment site and describe the production of lysates which transduce E. coli
material with high efficiency. Preliminary characterization of these transducing lysates is also presented.
MATERIALS A N D METHODS
Strains: Multiply-marked strains are listed in Table 1. Strains TR1854, TR1858, and TR1859 are deficient in DNA-restriction and modification systems; these strains were obtained from
C. COLSON, Institut Carnoy, Louvain, Belgium. Strain TR2270 was obtained from J. GOTS, Uni- versity of Pennsylvania, Philadelphia, Pennsylvania. E . coli strain TR367 was obtained from
A. J. CLARK of this department and it is identical to JC5459. The deletion mutation proAB47,
which is used extensively, is a large deletion which removes the gxu, proB, proA and a t a A
genes. (The ataA locus is the primary P22 prophage attachment site.) Mutant proA15 is a point mutant and proB25 a small deletion. All pro mutants and strains constructed from them were derived from Salmonella strain LT7 by M. DEMEREC and co-workers (MIYAKE and DEMERIC 1960; ITIKAWA and DEMEREC 1967). The F’ prolac episome is the F‘,,, described by Low (1972) and by Wu, JOHNSON and NEWTON (1970) and was obtained from J. GOTS, University of Penn- sylvania. The F’128 was derived from Hfr P804 (see Figure 4). P22 mutants were obtained from DAVID BOTSTEIN, Massachusetts Institute of Technology.
Media: The E medium of VOGEL and BONNER (1956) was used as minimal salts medium with 2% glucose as carbon source for most crosses. Whenever selection was made for growth on lactose as sole carbon source, the citrate-free E medium of BERKOWITZ et al. (1968) was used (NCE
lactose). Difco nutrient broth (0.2M i n NaC1) was used as maximally supplemented medium. Phage are stored in the T2 buffer of HERSHEY and CHASE (1952).
636 I. HOPPE A N D J. R O T H
TABLE 1 List of bacterial strains used
TR251
TA367 ( E . coli K12) TA1797 TR1810 TRI 825 TR1832 TR1837 TR1839 TRI 845
TR1846 ( E . coli K12) TRI 849 TR1852 TA1854 TR1857 TR1858 TR1859 TR2030 TR2031 TR2110 TR2246 TR2270 TR2277 TR2279 TA2288 TR2472 TR2473 TA2474 TR2612 TA2637 TR2638 TR2639 TR2654 TA2656
cysA1348 hid2527 sup-501 (amber)
s e r trp- B1- lacA74 strR su- (JC5469) arg1539 (isogenic with TA2030) strS proAB47/F pro+ lac+ strS proAB47/F' (P22) pro+ lac+ his644 (P22) (P22 prolacAz) his644 ( P a ) (P22 proflac+,,) proB25 (P22) (P22 pro,,)
proB25 (P22) (P22 proAl5)
s e r trp- B1- l a c strR/F' (P22) p r o f lac+ proAI5 (P22) (P22 proA1,)
proA15 (P22) (P22 proAzz)
rLTz r r L T 2 proCS0 (deficient in restriction and modification)
pyrB64 proAB47 (ataA-)
rS-m8- proCPU (deficient in restriction and modification) rLTZ m-LT2 r S m-s proC90 (deficient in restriction and
modification)
arg1539 proAB47/F' p ro f lac+ arg1539 proAB47/F' (P22) pro+ lac+
cysA1348 hid2527 sup-501 (amber) (P22 prolac,,) (P22 a d - 7 )
HfrB2 recA-l strR m t A 2 2 gxu-3 purE66
pyrB64 recA-1 strR
purB64 recA-1 strR proAB47 ( a t d - ) gxu proB25
arg1539 (P22) (P22 arg,) his644 (P22) (P22 pro+
his644 (P22) (P22 pro+ l a d A 7 , ) arg1539 (P22) (P22 arg,)
pyrB64 proAB47 (P22 prolac,,) (no helper phage)
pyrB64 proAB47 (P22 proZacA76) (no helper phage)
pyrB64 proAB47 (P22 prolac,,,) (no helper phage)
proA15 (P22 pro,,,) (no helper phage)
proB25 (P22 proAl5) (no helper phage)
the suspension was agitated during exposure. The irradiated cells were then aerated two hours a t 30" to permit lysis. Chloroform was added and the cultures were agitated for ten minutes. When large cultures were involved, phage was concentrated with polyethylene glycol (PEG)
by the method of YAMAMOTO et al. (1970). Cultures were made 0.5M in NaC1, and PEG was added to a final concentration of 10% (w/v). The cultures stood overnight at 4"; phage were then collected by continuous flow centrifugation. The pellets were resuspended in 0.85% NaCl (in preparation for CsCl gradients) or in T2 buffer (if phage was to be used directly in transduc- tion). The phage suspensions were cleaned by a low speed centrifugation (IO minutes a t 3,000G)
to remove debris and by a high speed centrifugation (50 minutes at 16,000G) to repellet the phage. Final phage stocks were prepared by resuspending these pellets i n saline (or T2 buffer). Since induction of P22 lysogens yields a high percentage of defective particles (ISRAEL 1967),
tails were added to many lysates by the method of ISRAEL, ANDERSON and LEVINE (1 967).
P22 SPECIALIZED T R A N S D U C TI O N 63 7
Episome transfer: All episome transfers were performed as spot tests on solid selective me- dium.
Isolation of g z u mutations: Mutants at the gzu locus such as strain TR2288 were selected as azaguanine-resistant mutants. Of 32 aLaguanine-resistant mutants, 6 proved to be linked to pro,
as described by GOTS, BENSON and SHUMAS (1972). The nature of the resistance of the other mutants is not known.
Construction of strains carrying the recA-I allele: Strain TR2246 (HfrB2, recA-I, strR, m e t A )
is a r e c strain constructed by BOTSTEIN and MATZ (1970). This Hfr was used to transfer a rec
mutation to a variety of strains; selection was made for s t r R [met+] and the recombinant progeny were scored for UV-sensitivity and recombination ability.
Nomenclature: Lysates containing specialized transducing phages have been numbered. For example, lysate A7 is obtained by UV-irradiation of strain TR1837; this lysate contains a par- ticular sort specialized transducing phage (designated P22 proZacA,) as well as plaque-forming P22 helper phage.
Construction of a strain carrying bath P22 lac,l, and P22 amN7 prophages: For experiments
on the requirement for helper phage, a specialized transducing lysate was needed, whose helper phage could not multiply during the course 3f a transduction. Since P22 lacA7 (as described later) lacks gene 20, this gene function presumably must be supplied by the helper phage. Therefore a strain was constructed which carries a gene 20 mutant, amN7, as helper prophage in addition to the P22 lac,, prophage.
This strain was made by transducing a n amber suppressor carrying strain (TR251) to LAC+. The transduction was done using a P22 lacA, lysate (which contains some P22 wild-type par- ticles) a t very low multiplicities (1 p.f.u./105 cells). Exogenous helper, amN7, was added at high multiplicity (5 p.f.u./cell). Most of the LAC+ transductants obtained proved to release phage which plate only on suppressor-carrying hosts. These transductants are presumed to carry the P22 l a A 7 prophage as well as a P22 amN7 prophage. One such transductant (TFUIIO) was used to prepare the needed lysate.
P22 anti-serum: The serum used is goat anti-serum prepared by Cappel Laboratories, Down- ingtown, Pa. Since the serum contains some antibody against Salmonella, it was first treated with heat-killed Salmonella cells. The precipitate was removed and the resulting serum was used in the experiments described.
RESULTS
1) A P22 attachment site
in
an E. coli F' prolac episomeSalmonella strains lacking the P22 prophage attachment site ( a t d ) are lyso- genized only rarely by P22. If such strains harbor an E. coli
F'
pro+ lac+ episome, lysogeny occurs readily. The lysogens formed carry the P22 prophage in associ- ation with the F' pro+ lac+ episome. These data are presented in Table 2. StrainTABLE 2
Insertion of P22 prophage into E. coli attachment site
Strain
Percentage of exposed. P22 t r i m a m attachment site Clones tested clones lysogenized LT2 Salmonella site present 96 88.5
proAB47 site missing 356 1.4
proAB47/F' pro+ lac+ E . coli site present 96. 54.2
638 I. HOPPE A N D J. R O T H
proAB47 carries a large deletion mutation which removes the proA and proB genes and also the prophage attachment site.
Two lines of evidence demonstrate that the lysogens formed in the proAB47/F’ pro lac strains are associated with the F’ element. Whenever lysogens lose the
F’
element they also lose the P22 prophage. Since Salmonella has no lac region of its own, cells losing the E. coli F’ pro+ lac+ episome can be identified as LAC- segregants. I n all cases, these segregant clones become simultaneously PRO- and phage-sensitive and do not release P22 phage. Apparently, loss of the episomal pro+ and lac+ genes is always associated with loss of the P22 prophage. When LAC+ clones were similarly tested, no prophage loss was detected. The second evidence for P22 insertion into theP
element is based on the conjugational trans- fer of the prophage from Salmonella into E. coli. The P22 attachment site seems to be a feature of P prolac and not of F’ episomes in general since no insertion into F’,,, could be demonstrated.2) Transfer of the P22 prophage into E. coli
The F’ pro+ lac+ episome carrying the P22 prophage can be transferred by conjugation into E. coli. This transfer probably results in a considerable amount of zygotic induction, but P22-lysogenic clones of E. coli can be isolated following F’ transfer. Since P22 phage particles cannot infect E . coli, the isolation of these P22 lysogens of E. coli is strong evidence that the prophage is associated with the
F“
pro+ lac+ episome.The Salmonella strain TR1825 [carrying the
P
pro+ lac+ (P22) episome] was mated with a lac- trp- sirR E. coli strain (TR367) and selection was made for LAC+ [St+] progeny which should arise by transfer of theF“
pro+ lac+ (P22) episome. Several progeny clones were picked, purified and UV-induced for phage preparation, Of nine clones tested, one released P22 phage. This strain is a merodiploid E. coli strain which segregates LAC-, P22-free clones. Table 3 presents assays of the P22 phage prepared following UV induction of this E. coli P22 lysogen (TR1846). The lysates have been assayed on a wild-type Salmonella strain and on Salmonella strains lacking one or both of the DNA-restriction systems known f o r Salmonella (COLSON and COLSON 1971; COLSON, COLSON and VAN PEL 1969, 1970). The results clearly indicate that the phage grown inTABLE 3
Assay of P22 grown in E. coli
Salmonella host strain Restriction systems present
Plating efficiency of P22 phage Grown in Grown in Salmonella’ E. coli K-12t
LT2 (wild type) Both S and LT2 systems 1.0 3.7 x 10-5
TR1858 Only LT2 system 0.8 1.5 x 10-4
TR1854 Only S system 0.8 0.2
TR1859 Neither system E l . 0 E l . 0
* Phage grmn lytically on S. typhimurium strain LT2.
P2 2 SPECIALIZED T R A N S D U C T I O N 639
E.
coli are restricted by Salmonella. Since the P22 lysogens of E. coli are sensitive to T4 and h phages, it seems that the P22 prophage does not interfere markedly with growth of these coliphages.3 ) Formation of proA, proB and lac high frequency transducing lysates
Specialized phages were formed by UV-induction of a Salmonella lysogen in which the P22 prophage is located adjacent to E. coli genetic material in an F’
episome. The lysogen used is a Salmonella strain (either TR1825 o r TR2031)
carrying a deletion mutation which removes the proA and proB and g x u genes as well as the P22 attachment site, a t d ; the strain harbors a n E . coli F’ prolac
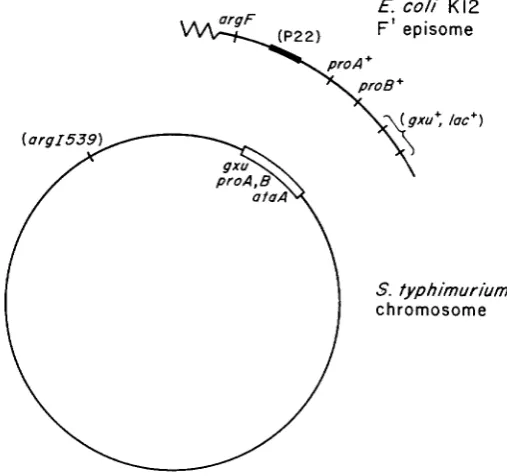
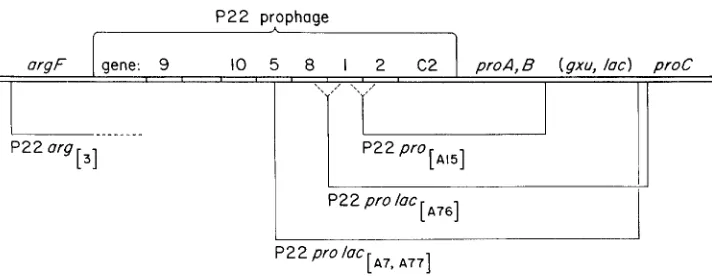
episome which carries a prophage attachment site. The P22 prophage is inserted into this E. coli attachment site and thus is located adjacent to E. coli genetic material. The structure of this strain is presented in Figure 1.
Phage grown lytically on a non-lysogenic strain of this genotype (e.g., TR1810
or TR2030), is unable to transduce proA, proB or lac genes to a Salmonella recipient by generalized transduction. This is true because the only proA and B
genes present are derived from E. coli which shows very poor homology with the S. typhimurium chromosome. The lac region is also derived from E. coli, but it is further unrelated in that Salmonella does not have a lac region. This lack of homology should not affect specialized transduction. If phages are formed in
E. coli K12
argF F’ episome
5- ‘02 2 )
FIGURE 1.-Genotype of the strain from which all specialized transducing phages were ob-
tained. The chromosome carries deletion proAB47 which removes the gz-u, proA and proB genes as well as the prophage attachment site aiaA. The P22 pro and P22 prolac phages were gen- erated following UV irradiation of strain TR1825, which is as diagrammed above except that no argl mutation is present. The P22 rrrgF phages were generated following UV irradiation of
640 I. HOPPE A N D J. ROTH
TABLE 4
Transduction of the E. coli lac region into Salmonella recipients
Recipient: his-644 ataA+ [lac-] *
Donor: proAB47/F’ proA+ proB++ lac+ (TR1810)
Type of lysate
Method of Phage plaque-forming units phage preparation required for one LAC+ transductant General transducing
LFT: low frequency HFT: high frequency
lytic growth of P22t UV induction of lysogen
UV induction of LAC
+
lysate (on TR1810) >lo11 (none detected)
transducing lysate (TRI 825) $’ 109
transducing lysate transductants$
TR1832 106 (A2)
TR1837 5 (A71
TR2473 2 (A76)
TR2474 3 (A77)
* The recipient is a his deletion mutant of Salmonella strain LT2; like the parent strain, this
t
The genotype of TR1810 is: proAB47/F’proA+ proB+ he+.$‘A P22 lysogen of TR1810 was prepared; this strain (TR1825) was TJV-induced to prepare
$ Data on four transductants are presented (A2, A7, A76 and A77). mutant is unable to ferment lactose.
phage.
which E. coli genes have become part of a specialized P22 transducing phage, then these genes should be inheritable by lysogenization.
Crosses involving transduction of the lac regio,n are presented in Table
4.
W e have never detected transduction of Zac by generalizod transduction using lyti- cally grown P22 (Table4,
line 1 ) . However, when strain TR1825 is UV-induced, the resulting lysate (LFT) transduces lac with low frequency (Table4).
This transducing ability is presumably due to the presence of specialized transducing phages ( P 2 2 lac) formed by improper prophage excision.High frequency transducing lysates (HFT) were obtained by UV-induction of the LAC+ transductants formed by the LFT lysate, These transductants were UV-induced and the resulting lysates used in transduction. The transducing frequency observed varies from one transductant to the next. Lysates derived from several of these transductants (A2, A7, A76, A77) are presented in line 3 of Table
4.
By a similar series of transductional crosses, high frequency transducing lysates were prepared for the proA and proB genes. Phage prepared by UV-induc- tion of TR1825 (cf. Figure 1) was used in crosses with proA15 and proB25 recipients. The pro+ recombinants were picked and purified; lysates prepared following UV-induction were used in a search for high frequency transducing lysates. Results typical of these crosses are presented in Table 5 . In these crosses,
P22 SPECIALIZED TRANSDUCTION
TABLE 5
Transduction of E . coli proA and proB genes into Salmorzella Donor: TR18 10 (proAB47/F’pro+ lac+ )
641
Preparation of donor phage
~~~~ ~~ ~~~
Phage plaque-forming units required for one PRO+ transductant nsiug various recipient strains
proAB47 proAl5 proB25 (no P22 attachment site)
lytic growth (on TR1810) 3.3 x 10s 6.2 x 108 >i.o x 109 lysogen (TR1825) * 1.0 x 107 1.5
x
107 1.9x
10;sUV induction of TJV induction of PRO f
transductants TRl85at 3.2 x 1013 4.6
x
103 1.5x
104 TR1849-t 2.1x
IO3 1.5x
1013 7.1 x 104 TR1839$ 4.5x
103 3.6 x 103 1.9x
1016 TR1845$ 3.8 x 102 2.8 X IO2 9.1x
105* Strain TR1825 is a P22 lysogen of TR1810, in which the prophage is inserted into the E. coli
F’ pro+ lac+ episome.
t
This transductant was selected as a PRO+ recombinant using proAI5 as recipient and donor phage prepared by UV induction of TR1825. The transductant was UV-induced to yield the lysate used in the above crosses.$ This transductant was selected as a PRO+ recambinant using proB25 as recipient and donor phage prepared by UV induction of TR1825. The transductant was UV-induced to yield the lysate used in the above crosses.
duced during the lytic cycle, as described by SMITH-KEARY (1966) and by
JESSOP (1972).
4) Construction of specialized phages carrying the argF genes
E. coZi K12 has two distinct genes for ornithine transcarbamylase (OTCase) (GLANSDORFF, SAND and VERHOEF 1967; LEGRAIN et al. 1972). These genes are argZ, mapping at minute 85 on the chromosome, and argF, mapping at minute 7
near the pro and lac genes. Salmonella has only the argZ gene and lacks a locus homologous to the argF gene of E . coli (SYVANEN and
ROTH
1972). Since the E. coli argF gene is located near the P22 prophage attachment site on theF’
pro+ lac+ episome, attempts were made to construct a P22 argF transducing phage.
This was done by basically the same method used f o r formation of P22 lac and P22 pro phages. The only difference is that all strains used carried a chromosomal argZ mutation. The donor is Salmonella strain TR2031 (argZ539 proAB47/F’ arg+ (P22) prof lac+) whose F’ episome is derived from E. coli K12. This is the same F’ episome used to construct the P22 pro and P22 prolac phages.
Lysates of this strain were prepared following UV-induction. The only func- tional OTCase gene in this strain is the E. coli argF gene on the episome. The recipient is a Salmonella mutant argZ539. In the transduction, selection is made for ARG+ (OTCase+) recombinants. These recombinants will almost never form by generalized transduction since the only functional OTCase gene carried by the donor is the E. coli argF locus. The E. coli-Salmonella difference plus the lack
642 I. HOPPE A N D J. ROTH
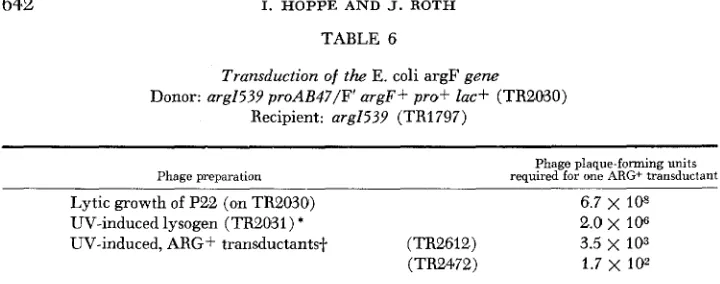
TABLE 6
Transduction of the E. coli argF gene
Donor: arg1539 proAB47/F' argF+ prof ZQC+ (TR2030) Recipient: arg1539 (TR1797)
~
Phage preparation required for one Phage plaque-forming units ARG+ transductant
Lytic growth of P22 (on TR2030) 6.7 x IO8
UV-induced lysogen ( ~ ~ 2 0 3 1 ) * 2.0
x
106UV-induced, ARG+ transductantsf. (TR2612) 3.5
x
103(TR2472) 1.7 x lo2
* Strain TR2031 is a P22 lysogen of TR2030; its genotype is: mg1539 proAB47/F' argF+ (P22) prof lac+.
+
Strains TR2612 and TR2472 are ARG+ transductants obtained using recipient strains arg1539 and donor phage prepared by LJV induction of TR2031 (cf. line 2 in above). The genotype of TR2612 and TR2472 is presumed to be arg1539 (P22) (P22 arg).ever, specialized transduction of argF should not be impaired by these strain differences.
ARG+ transductants were picked and purified and were then UV-induced to produce potential high frequency transducing lysates. Typical results are pre- sented in Table 6.
5 ) Evidence for Specialized Transduction
Three lines of evidence suggest that the high frequency transducing lysates described above contain specialized transducing phage.
A) The transductants are unstable. All of above phages (P22 lac, P22 pro,
P22 arg) give transductant clones in which cells of the original phenotype appear.
For example, when a LAC+ transductant clone (obtained using P22 lacA7) is grown overnight in non-selective medium, 1-10% of the cells in the final popu- lation are found to be LAC-. Similar segregant frequencies are found for trans- ductants obtained using the P22 arg and P22 pro transducing phages. This demonstrates that the transduced genes are not stably integrated. All such segre- gants are lysogenic; this suggests that they have lost their transducing-phage genome and retained a helper prophage.
B) The recipient strain must have a prophage attachment site for optimal transduction frequency. In Table 7 (line 3 ) it can be seen that all specialized transducing phages give greatly reduced transduction frequencies when the recipient strain carries mutation proAB47, a deletion which removes the P22 attachment site (ataA)
.
This deletion does not affect generalized transduction. The pyrB region (last column of Table 7 ) was used to demonstrate this since theLac region cannot be transduced by generalized transduction.
C) Transduction by these phages does not show absolute dependence on a
P22 SPECIALIZED T R A N S D U C T I O N
TABLE 7
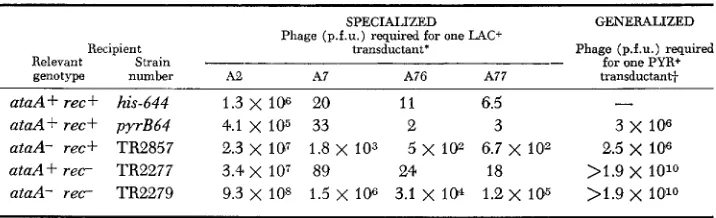
Role of recA and ataA in generalized and specialized transduction
643
Recipient Relevant Strain genotype number ataA + rec f his-644 ataA+ rec+ pyrB64 ataA- rec+ TR2857 a t a A f r e c TR2277 aiaA- r e c TR2279
SPECIALIZED Phage (p.f.u.) required for one LAC+
transductant*
A2 A7 A76 A77
1.3
x
106 20 1 1 6.54.1 x 105 33 2 3
3 . 4 x 107 89 24 18 2.3 x 107 1.8 x 103 5 x 102 6.7 x io2
9.3 x 108 1.5
x
106 3.1 x 104 1.2 x 105GENERALIZED
Phage (p.f.u.) required for one PYR+ transductant+
-
3x 106
2.5 x IO6 >1.9 x 1010 >1.9 x 1010* A2, A7, A76, A77 are various specialized P22 prolac transducing lysates.
+
Donor P22 int-4 grown lytically on LT-2 wild-type Salmonella.It should be noted, however, that the recA mutation does cause some reduction in frequency of transduction. This may reflect some contribution of recA to circularization or insertion of the transducing phage. The reduction could also be due, at least in part, to the decreased viability of transductant clones carrying the recA mutation.
Very low recombination frequencies were observed when the recipient strain has both an attachment site (ataA) deletion and a recA mutation. The few transductants which are independent of both crtaA+ and recA+ may represent int-mediated lysogenization at secondary prophage attachment sites or insertion at random sites by the phage’s generalized recombination system.
6) Helper phage is required for optimal transduction by P22 lacAr
I t was our expectatioa that the high frequency transducing lysates would contain both defective specialized transducing phage and wild-type P22 particles.
If
this helper phage is required for transduction, then the efficiency of transduc- tion should decrease when low multiplicities of infection are used (< 1 .O).
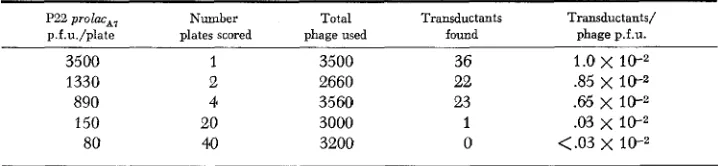
Thisproved difficult to check since helper phage can multiply on the transduction plate and soon raises the true multiplicity of infection. The following experiment was done using extremely low multiplicities. So few phage particles were placed on each plate that different phage-infected cells were spatially isolated. A total of approximately 3,000 phage particles of a P22 lacA7 lysate were distributed to several lawns of a lac- recipient strain. The number of p.f.u. plated varied from 150/plate to 3,50O/plate. The reduction in transducing efficiency is apparent once helper phage and transducing phage are prevented from infecting the same cell
(Table 8).
The helping effect of wild-type P22 is also shown by using exogenously sup- plied helper. A lysogen was constructed which carries P22 prolac,, and P22 amber mutant amN-7 as helper (see MATERIALS AND METHODS). When a lysate
644 I. HOPPE A N D J. ROTH
TABLE 8
Helper requirement of P22 prolac*, for optimal transduction e f f i c i e q
P22 prolac,, Number Total Transductants Transductants/
p.f.u./plate plates scored phage used found phage p.f.u.
3500 1 3500 36 1.0
x
1 0 - 21330 2 2660 22 3 5
x
10;2890 4 3560 23 .65 x 10-2
150 20 3000 1 .03 x
80 40 3200 0 <.03
x
l0rzIn each cross, recipient lawn was 2 x 108 cells of the LAC- strain, his 644. Selection was made
for LAC+ transductants on NCE medium containing lactose as sole carbon source and supple- mented with histidine. Donor phage was prepared by UV induction of strain TR1837.
TABLE 9
Demonstration of helper effect using (P22 amN7) (Pa2 prolmft,) domr phage
Donor phage (p.f.u.) used per 3 X lo8 cells
Number of L4C+ transductants ~~
Recipient with Recipient without amber suppressor amber suppressor (his-644)
(TR251)
-
Helper+
Helper 7 x 1067 x 105 7 x 104 7 x 103 7 x IO'
70 7
~
confluent
-
1,030 8,000 -
340 108
-
21 7 0 1,724
6 0 375
0 0 24
0 0 2
-
10,000Cells and phage were mixed in nutrient broth and incubated 20 min for adsorption before plating on selective medium. Donor phage was prepared by UV induction of strain TR2110. Helper phage is P22 amber mutant a d 7 grown on strain TR251, used at a multiplicity of
3 phage/cell; helper phage alone shows no transducing ability. Recipient strain his-644 carries no amber suppressor; thus phage multiplication should not occur in the course of the experiment. In the donor phage lysate, a d 7 revertants are present at about 1/1W p.f.u.; these revertants can multiply and probably act as helper to permit the LAC+ transductants seen a t high donor phage levels.
Table 9 ) . Addition of pure P22 amN7 phage to the transducing lysate greatly increases the transduction efficiency under these conditions.
7) The specialized transducing phages are defective
P22 SPECIALIZED TRANSDUCTION
[ ~ 1 5 1
[ ~ 7 6 1
P22 arg [ 3 1
’
P22 proI
P2 2 pro lac
645
proc
FIGURE 2 . 4 t r u c t u r e of specialized transducing phage. The genetic map presented above is that o€ the E . coli K12 F‘ proluc genetic material €rom which these phages are derived. Solid horizontal lines below the genetic map indicate the extent of this material which is contained in the several specialized phages. Uncertainties are indicated by dotted lines.
Helper-free transductants were first sought by performing transduction at extremely low multiplicities of infection. This method was successful for P22 proAls and P22 argIs1; transductants were obtained which are phage-sensitive and release no viable P22 phage.
I n the case of P22 proZacA7, low multiplicity of infection did not serve to permit isolation of helper-free transductants. Apparently, wild-type P22 is required as a helper for this transducing phage (see above), and this helper phage usually lysogenizes the transduced cell. I n order to reduce the probability of lyosgeny of transductants by helper, a recipient strain was used which lacks the prophage attachment site. In this strain, transducing phage and helper must insert at secondary attachment sites and thus insert with reduced frequency. This seems to increase the probability of obtaining transductant clones which lack the helper phage. Using this recipient, several transductant clones were isolated which are sensitive to P22.
All of the above helper-free, P22-sensitive, transductants were tested f o r their content of P22 phage genes. This was done by checking the transductants ability to support the growth olf a series of P22 amber mutants. Results are presented in Table 10. The P22 mutants will grow only if the transductant cell can provide the missing phage function. This method does not require expression of the pro- phage genome since recombination between prophage and the mutant phage genome generates wild-type recombinants which form plaques (see CHAN and BOTSTEIN 1972). As seen in Table IO, phages P22 Zac,,, P22 lacA76, P22 lacA77 and P22 IO^^^ all contain part of the P22 genome. The P22 argIsl transductants showed no detectable P22 genes by this test.
8) Relatedness of P22 and P22 lacA7
P22 SPECIALIZED TRANSDUCTION 64 7
dence indicate that P22 laca7, best characterized of our phages, is derived from P22. Most of these criteria apply to the P22 pro and P22 m g phages as well.
a ) The transducing phage seems to require the P22 prophage attachment site since transduction is greatly reduced when the recipient lacks this site
(Table 7 ) .
b) The transducing phages are defective but include a series of genes which can supply missing functions to P22 mutants (Table 10).
C) When wild-type P22 is produced by UV-induction of a lysogen, a high proportion of the phage particles released are defective in tail structures
(ISRAEL 1967). Tails can be added to such defective P22 particles to make them infective (ISRAEL, ANDERSON and LEVINE 1967). Addition of P22 tails to transducing lysates of P22 lacA7 causes a n increase in plaque- forming units and also a n increase in transducing activity. Thus the trans- ducing particles, like P22, are deficient i n tail fibers, and, like P22, these defective particles accept P22 tails.
d) A transducing lysate of P22 lacA7, to which P22 tail fibers had been added to correct defective particles, was treated with antiserum against wild-type P22. The plaque-forming activity and transducing activity of the lysate were inactivated at the same rate. (Transducing activity was assayed in the presence of added helper to be sure that the drop in transducing activity was not due to loss of the required helper.)
I n contrast to the above lines of evidence, one finding suggests that P22 and P22 ZmA7 might not be related. When a P22 lacA7 transducing lysate, to which tails have not been added, is treated with P22 antiserum, plaque-forming activity
in inactivated a t a normal rate, but transducing activity is inactivated very poorly. This suggests a n immunological difference between the infective P22
lacn7 phages and the P22 helper phage present in the lysate before tail addition.
Perhaps a small fraction of the total transducing phage genomes is packaged in
the heads of an unrelated phage; before tail addition, when most P22 particles are tail defective, this small fraction would represent a major proportion of the functional transducing particles. This explanation (courtesy of D. BOTSTEIN)
accounts for our finding but is not yet supported by direct evidence. All lysates described above were prepared by UV-irradiation of strain TR1837 which is derived from strain LT7.
The failure of P22 antiserum to inactivate transducing activity is similar to a n observation of BENZINGER (1962). BENZINGER found that this generalized trans- ducing activity of P22 phage released spontaneously by P22 lysogens of strain LT2 was not inactivated by P22 antiserum. This, like our observations for P22
lac, may be due to packaging of genetic material into heads of a phage that is immunologically unrelated to P22. The observations may also be a consequence of the tail defects of P22 phage released following UV induction. Spontaneously released P22 is also extremely defective in tails; when culture fluid of a P22
648 I. HOPPE A N D J. R O T H
stantially tail-defective particles were able to inject transducing DNA, this would account for all the above observations without involvement of a n unrelated phage.
9) Density of transducing particles
The density of P22 specialized transducing phage was checked to see if they differ in DNA content from wild-type P22 phage. Since P22 is thought to package its DNA by “headful measuring”, one would expect the DNA content of trans- ducing phages to be identical to wild-type P22. This is in contrast to phages such as X which recognize sequences in determining the amount of DNA packaged.
In
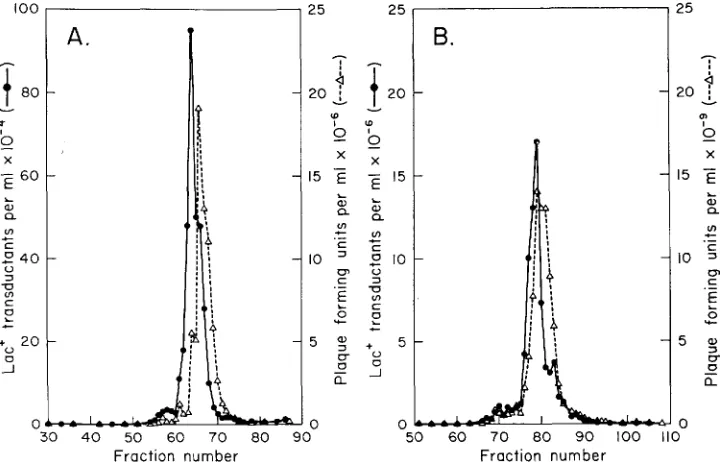
the latter sort of phage considerable leeway in DNA content is permitted and specialized phages can differ considerably from wild type in their density.Figure 3 presents CsCl density gradients of transducing lysates of P22 prolacA,. Transducing activity was assayed by transductional crosses with a lac- Salmonella recipient (his-644). In these crosses a n excess of helper phage (P22 mutant, amN7) was provided. Plaque-forming activity of the P22 prolac lysate was measured by plaque assay of each fraction. Panel A presents data for a lysate to which no tails have been added; panel B presents data for the same amount of the same lysate but with tails added before centrifugation. Peaks of transducing
100
4
80-
P I
0 6 0 L a, Q ul c c
e
4 03
-U ul
c
z
L
+ 20
-I
0
U
B.
I
0 0
25
-
4
20 ;
0 v m I X 15 L Q) a U) c 0, c c .- 10 = . _
E
5 s 0 c U 0-
a nFraction number Fraction number
P22 SPECIALIZED TRANSDUCTION 649
activity and plaque-forming activity coincide closely, but the transducing peaks consistently appear somewhat denser than plaque-forming units. This slight difference is probably due to differences i n base composition of DNA’s of the two phage types.
10) Gena cwried
b./
various P22 specialized phagesThree types of phages have been constructed: those selected for transduction of lac, of proA (or proB)
,
and those selected for transduction of argF genes. Each of these phages was tested f o r the presence of other genes known to be in this region. Results are presented diagrammatically in Figure 2.All phages carrying lac also carry proA and proB. All phages selected for transduction of one pro gene ( A or B ) also carry the other pro gene; none of these P22 pro transducing phages carries lac. Neither P22 pro nor P22 Zac phages carries argF. None of the argF phages tested carries either pro or lac. None of the P22 prolac phages includes the proC gene.
11) A phage which carries the gene (gxu) for guanine-xanthine phosphoribosyl
transferase
It
has recently been reported (GOTS, BENSON and SHUMAS 1972; CHAN andMARTIN
1972) that the gene (gxu) for guanine-xanthine phosphoribosyl trans- ferase is located very close to the proA and proB genes of S. typhimurium. W e therefore checked several of the P22 specialized phages fomr the presence of thegnu gene. The gxu gene is carried by the P22 lac phages but not by a P22 pro
transducing phage.
I n performing these tests a strain was constructed which carries a gxu muta- tion and a proB25 mutation (TR2288). The double mutant was selected as an azaguanine-resistant derivative of mutant proB25. The azaguanine-resistance mutation is linked to proB25, as reported by GOTS, BENSON and SHUMAS (1972).
This strain was used transduction crosses With P22 p i O A 1 5 , P22 proAI9 and with
P22 lac,, b h frequency transducing lysates. Selection was made for PRO+ and
PRO+ LAC+, respectively. The transductant clones were then checked for
azaguanine resistance. Transductants using P22 pro phage were still azaguanine- resistant. Transductants using P22 lacA7 phage have become simultaneously
PRO+, LAC+ and azaguanine-sensitive. This property was associated with the specialized transducing phage, since LAC- segregants which are presumed to have lost the phage becomes simultaneously LAC-, PRO- and azaguanine- resistant.
The P22 lac,, phage was tested in another way. A double mutant strain gzu
purE66 was obtained from J. GOTS. The gxu mutation renders this strain unable
to use guanine or guanosine as a purine source; only adenine or adenosine can satisfy the purine requirement. This strain was transduced with P22 lacA7,
selecting LAC+ transductants. The transductants obtained had become gxu+ in that they had gained the ability to utilize guanine and guanosine as a purine source. LAC- segregants lost both the ability to ferment lactose and the ability to utilize guanine and guanosine.
650 I. HOPPE A N D J. ROTH
F’,28 pro lac
__-_
; gxu prolAEl ataA argF
-
lac I proC ,Parental E c o l i
-
chromosome Hfr
---_
gxu proB,A atoA proC
S. typhimurium
_---
chromosome
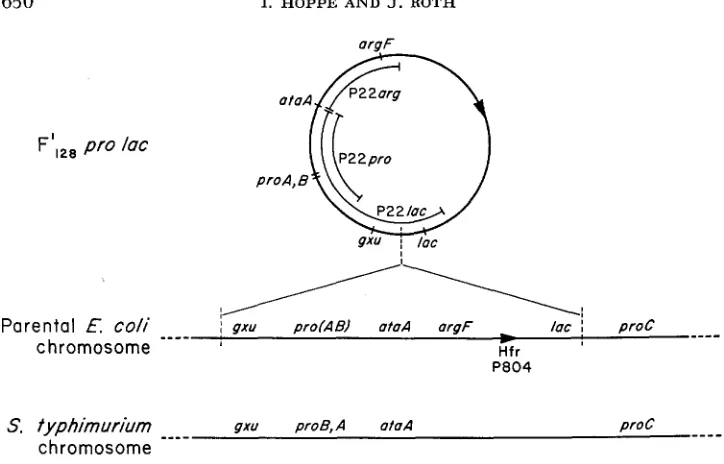
FIGURE 4.-Suggested map of E. coli prolac region. At the top of the figure is the map of the
prolac episome deduced from the composition of P22 specialized phages. The extent of ma- terial included in the three types of transducing phages is indicated inside the circular map. Be- low the F’ map is a suggested map of the prc-lac region of P804, the Hfr from which F’,,, prolac
is derived. The transfer origin is as described by TAYLOR and TROTTER (1972). The g z u gene posi- tion is based only on homology with the map order in S. fyphimurium, presented a t the bottom of the figure. The Salmonella Chromosome has no genes homologous with argF and lac.
The exact map of the prc-lac region of E. coli is uncertain. The problems that have arisen in mapping this region have been discussed by TAYLOR and TROTTER
(1972). The P22 specialized phages described in this paper are derived from an E. coli F’ episome carrying this region and thus may shed light on gene order in the E . coli chromosome.
Figure 4 (top) presents the probable structure of the F’ p r d a c episome. The position of the ataA and gxu regions is based on data presented in Figure 2; the order of gzu and lac is uncertain. In the middle of Figure 4 is a suggested map for the chromosome of P804, the Hfr from which F’ prolac was derived. The position of the Hfr origin is from Low (1972). The gzu gene is placed at the left (rather than near lac)
,
based only on homology with the position of the gzu gene in Salmonella (GOTS, BENSON and SHUMAS 1972). If this order is correct, it suggests that the lac region was brought close to the g x u gene by the events which led to formation of F‘ prolac.P22 SPECIALIZED TRANSDUCTION 65 1
soma1 map. If the episome has undergone inversion or rearrangement since leaving the E . coli chromosome, then o w conclusions would be invalid.
DISCUSSION
Specialized transducting phages derived from P22 potentially show two unique features as a consequence of P22’s “headful measuring” mode of DNA packaging.
If the abnormal prophage excision events generate a specialized phage genome that is larger than a normal P22 phage genome, the transduced fragment will have no terminal redundancy and will be unable to circularize. Thus more than one phage particle might be required to carry the entire genome. A situation like this has been described by WATANABE et al. (1972) and by CHAN et al. (1972) for their P22 transducing phage which carries determinants f o r tetracycline resistance. Another case of this may be the P22 pro phages described by JESSOP
( 1972).
A second possible consequence of the P22 packaging mechanism would be realized if the specialized transducing phage were shorter than a P22 genome.
In this case a single transducing phage particle might include a genome with a terminal redundancy. In the case of a n extremely small transducing phage genome, several copies of the genome might be required to make u p a “headful”. Transductants formed by such phage might carry a duplication of the transduced material.
The above considerations may make P22 a particularly versatile virus f o r construction of specialized transducing phage since there may be few restrictions on the size of the transducing phage genome. On the one hand, transducing phage may maintain the whole phage genome and carry added bacterial genes (as in the case of the P22 tetR phage). On the other hand, the transducing particle might contain virtually no phage genes; the minimum requirement might be one attachment site or a small bit of homology to permit insertion into the chromo- some with the aid of a helper phage. A situation like this may be represented by the P22 arg phage described here o r by the P22 pyrB phage described in the accompanying paper
(KAYE
et al. 1973). We have been unable to detect phage genes associated with either of these two transducing phages.We do not know the exact genome structure or mode of replication of the P22
prolac phage (A7) described here. If the transducing phage and the helper joined to form a single replicated genome, as seems to be the case for the P22 tetR phage, then one would expect that P22’s “headful measuring” mechanism might generate a variety of defective phage genomes which are portions of this helper- transducing phage complex. This was found for P22 tetR, (CHAN et al. 1972). I n the case of P22 prolac lysates, however, there is no evidence for this; plaque- forming ability drops linearly with dilution, Apparently, the plaque-forming ability of P22 prolac lysogens is due to fully competent single particles.
652 I. HOPPE A N D J. R O T H
present in a P22 lysate, that lysate should show a higher titer of plaque-forming units when plated on a n L phage lysogen. P22 tetR lysates show a 1000-fold greater efficiency of plating on an L phage lysogen than on a standard LT2 indi- cator (CHAN et al. 1972). In contrast to this situation, P22 prolac phages show the same efficiency of plating on a n
L
lysogen as on a non-lysogenic indicator strain. This suggests that the P22 prolac lysates do not contain a large number of defective genomes; apparently the defective transducing phage particles which are present either do not form plaques on an L phage lysogen or are present at such a low frequency ( < l o % ) that they do not make a detectable contribution to plating efficiency on theL
phage lysogen.Our current hypothesis is that P22 prolac and P22 helper prophage are located in tandem in the chromosome and replicate as a unit upon induction. We propose that the P22 prolac genome is very close tu the size of the P22 genome. Therefore, when the concatenate is packaged, the “headful measuring” mechanism can operate to form two sorts of phages, (1) P22 wild-type particles with a normal permuted terminal redundancy, and (2) P22 prolac transducing particles which may be able to circularize independently if the P22 prolac genome is equal to or slightly less than the size af P22. If P22 prolac is slightly larger than the P22
genome, then the transducing phage genomes would not contain a terminal redundancy. The helper requirement for transduction might be due to the need for aid in circularization of P22 prolac. This hypothesis is made more likely by the recent finding of TYE, HUBERMAN and BOTSTEIN (1974) that the circular permutations of the P22 genome are not random; only about 20% of the P22
genome is represented i n the permuted terminolagy as redundant material. An alternative to this model is that P22 prolac and the P22 helper phage repli- cate as physically independent entities. If this is the case then normal P22 phages would be packaged from the helper concatenate and P22 prolac phages would be packaged from the second concatenate. Depending on the genome size of P22
prolac, these transducing phage chromosomes might or might not contain their
own terminal redundancy.
The work described here involves only transduction of E. coli genetic material derived from F’ episomes. This material was used as a means of reducing the background level of general transduction activity. However, since i t is now clear that P22 can form specialized transducing phages, it should be possible to con- struct phages carrying pieces of the Salmonella chromosome. In order to do this, generalized transduction can probably be restricted by use of rec- recipients or through use of recipients carrying large deletions of the genes of interest.
Note added in proof:
Charles Miller, Case Western Reserve Medical School, has found that phage
P22 prolac,, carries a n additional gene, pepD. The pepD gene codes for a dipepti- dase and is closely linked to both the gxu and proB loci in Salmonella (cf. Fig. 4).
P2 2 SPECIALIZED T R A N S D U C T I O N 653
We thank DAVID BOTSTEIN for helpful advice, critical discussion and numerous phage mu-
tants. JOANNE UOMINI, R. P. ANDERSON, J. BARRAVECCHIO, B. RATZKIN and G. ROBERTS made helpful comments during preparation of the manuscripts. This work was supported by Public Health Research Grant GM 18633 from the National Institute of General Medical Science.
L I T E R A T U R E C I T E D
BENZINGER, R., 1962
BERKOWITZ, D. J. HUSHON, H. J. WHITFIELD, JR., J. ROTH and B. N. AMES, 1968 BOTSTEIN, D., 1968
BOTSTEIN, D. and M. LEVINE, 1968a
Phages released from S. typhimurium lysogenized by P22. Virology 18: Procedures w3-645.
for identifying nonsense mutations. J. Bacteriol. 96: 215-270. mediates. J. Mol. Biol. 34: 621-641.
Synthesis and maturation of Phage P22 DNA. I. Identification of inter- Synthesis and maturation of P22 DNA. 11. Properties of temperature-sensitive phage mutants defective in DNA metabolism. J. Mol. Biol. 34: 643- 654.
-,
196813 Intermediates in the synthesis of phage P22 DNA. Cold Spring Harbor Symp. Quant. Biol. 33: 659-667.BOTSTEIN, D. and M. J. MATZ, 1970 A recombination function essential to the growth of bac- teriophage P22. J. Mol. 54: 41 7-440.
CAMPBELL, A., 1971 Genetic Structure in The Bacteriophage Lambda, pp. 13-44. Edited by
A. D. HERSHEY. Cold Spring Harbor Laboratory.
CHAN, J. Y. and R. G. MARTIN, 1972 Purine Phosphoribosyltransferases of Salmonella typhi- murium. J. Bacteriol. 112: 1010-1013.
CHAN, R. K. and D. BOTSTEIN, 1972 Genetics of Bacteriophage P22. I. Isolation of prophage deletions which affect immunity to superinfection. Virology 49: 257-267.
CHAN, R. K., D. BOTSTEIN, T. WATANABE and Y. OGATA, 1972 Specialized transduction of tetra- cycline resistance by Phage P22 i n Salmonella typhimurium. 11. Properties of a high- frequency-transducing lysate. Virology 50 : 883-898.
A new Salmonella typhimurium DNA host specificity. J. Gen. Microbiol. 69: 345-351.
Host controlled restriction mutants of Salmonella typhimurium. J. Gen. Microbiol. 5 8 : 57-64. -, 1970 Chromosomal loca- tion of host specificity in Salmonella typhimurium. J. Gen. Microbiol. 60: 265-271.
Genetics of plasmids in Salmonella typhimurium. Nature 204: 1112-1113.
The dual genetic control of ornithine trans- carbamylase synthesis in Escherichia coli K12. Mutation Res. 4: 743-751.
Genetic separation of hypoxanthine and guanine-xanthine phosphoribosyltransferase activities by deletion mutations in Salmonella typhimurium. J. Bacteriol. 112: 910-916.
HERSHEY, A. D. and M. CHASE, 1952 Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36: 38-56.
ISRAEL, V., 1967 The production of inactive P22 particles following induction. Virology 33:
31 7-322.
ISRAEL, J. V., T. F. ANDERSON and M. LEVINE, 1967 In vitro morphogenesis of phage P22 from heads and base plate parts. Proc. Nat’l. Acad. Sci. U.S. 57: 286291.
ITIKAWA, M. and M. DEMEREC, 1967 Salmonella typhimurium proline mutants. Genetics 95:
1189.
COLSON, C. and A. M. COLSON, 1971
COLSON, A. M., C. COLSON and A. VAN PEL, 1969
DUBNAU, E. and B. A. D. STOCKER, 1964
GLANSDORFF, N., G. SAND and C. VERHOEF, 1967
654 I. HOPPE A N D J. ROTH
JESSOP, ADRIENNE, 1972 A specialized transducing phage of P22 for which the ability to form plaques is associated with transduction of the proAB region. Molec. Gen. Genetics 114: KAYE, R., J. BARRAVECCHIO and J. ROTH, 1974 Isolation of P22 specialized transducing phage
following F'-episome fusion. Genetics (this issue).
LEGRAIN, C., P. HALLEUX, V. STALON, N. GLANSDORFF, 1972 The dual genetic control of ornithine carbamylasetransferase in Escherichia coli-a case of bacterial hybrid enzymes. Eur. J. Biochem. 27: 93-102.
Escherichia coli K12 F-prime factors, old and new. Bacteriol. Rev. 36: 587-607.
Proline mutants of Salmonella typhimurium. Genetics 45:
755-762. 2 14-222.
Low, K. BROOKS, 1972
MIYAKE, T. and M. DEMEREC, 1960
OZEKI, H. and H. IKEDA, 1968
RHOADES, M., L. A. MACHATTIE and C. A. THOMAS, 1968 The P22 bacteriophage DNA mole- cule. I. The mature form. J. Mol. Biol. 37: 21-40.
SMITH-KEARY, P. F., 1966 Restricted transduction by bacteriophage P22 i n Salmonella ty- phimurium. Genet. Res. 8 : 73-82.
SYVANEN, J. M. and J. R. ROTH, 1972 Structural genes for ornithine transcarbamylase i n Sal- monella typhimurium and Escherichia coli K-12. J. Bacteriol. 110: 66-70.
TAYLOR, A. L. and C. D. TROTTER, 1972 Linkage map of Escherichia coli strain K-12. Bacteriol. Review 36 : 504-524.
TYE, B.-K., J. A. HUBERMAN and D. BOTSTEIN, 1974 Non-random circular permutation of phage P22 DNA. J. Mol. Biol. (In press.)
VOGEL, H. J. and D. M. BONNER, 1956 Acetylornithinase of Escherichia coli: partial punfica- tion and some properties. J. Biol. Chem. 218: 97-106.
WATANABE, T., Y. OGATA, R. K. CHAN and D. BOTSTEIN, 1972 Specialized transduction of tetra- cycline resistance by phage P22 in Salmonella typhimurium. I. Transduction of R factor 222 by Phage P22. Virdogy 50: 874-882.
Transduction by phage P22 in a recombination-deficient mutant
of Salmonella typhimurium. Virology 36: 271-276.
Operon coordination in different bac- terial hosts. J. Bacteriol. 103: 318-322.
Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to
large-scale virus purification. Virology 40: 734-744.
Corresponding editor: D. KAISER Transduction mechanisms. Ann. Rev. of Genetics 2: 245-278.
WING, JENNIFER PATAI, 1968
W u , Po CHI, P. F. JOHNSON and AUSTIN NEWTON, 1970