0095-1137/95/$04.0010
Copyrightq1995, American Society for Microbiology
Biochemical Fingerprinting Compared with Ribotyping and
Pulsed-Field Gel Electrophoresis of DNA for
Epidemiological Typing of Enterococci
INGER KU¨ HN,1* LARS G. BURMAN,2SARA HÆGGMAN,2KJELL TULLUS,3 ANDBARBARA E. MURRAY4
Microbiology and Tumorbiology Centre, Karolinska Institute, S-171 77 Stockholm,1Swedish Institute for
Infectious Disease Control, S-105 21 Stockholm,2and Department of Paediatrics, St Go¨ran’s
Children’s Hospital, S-112 81 Stockholm,3Sweden, and Center for Infectious
Diseases, University of Texas Medical School, Houston, Texas 770304
Received 2 March 1995/Returned for modification 16 May 1995/Accepted 3 August 1995
The Phene Plate (PhP) biochemical fingerprinting system for bacteria is based on measurements of the kinetics of bacterial biochemical reactions. This system was modified for typing of enterococci and was compared with DNA typing by pulsed-field gel electrophoresis and with ribotyping by using 45Enterococcus
faecalisisolates from international collections. It was also used to study 170 fecal enterococcal isolates from
healthy individuals and 28 isolates ofE. faecalisfrom the blood of neonates. The PhP system showed a high degree of discriminatory power for unrelated enterococcal isolates. Among the 170 unrelated fecal isolates, 107 PhP types were found, and the diversity index was 0.97. DNA typing discriminated 27 types among the 45 isolates from international collections, PhP typing discriminated 19 types, and ribotyping discriminated 5 types. In most cases, when isolates were of the same DNA type, they were also of the same PhP type, and the level of agreement between these two methods was high (96%). A combination of PhP typing and DNA typing identified 34 different types, but ribotyping did not yield any further discrimination. PhP typing ofE. faecalis
isolates from healthy individuals (n589) and from the blood of neonates with septicemia (n528) yielded a diversity of 0.93 for both populations and similar major PhP types in both populations. Thus, the isolates from blood seemed to consist of a normalE. faecalispopulation, without a dominance of certain strains associated with virulence. We conclude that the PhP system is useful for epidemiological studies of enterococcal isolates, yielding results similar to those obtained with DNA typing by pulsed-field gel electrophoresis. Since PhP typing is a method that is simple and rapid and that is based on automatic evaluation of the data, it is suitable for analyzing large numbers of isolates and can be used alone or in combination with DNA typing for epidemio-logical and ecoepidemio-logical studies of enterococci.
Enterococci are members of the normal intestinal flora of humans and animals and can be widely found in plants, soil, and polluted water (1). They are increasingly common causes of nosocomial infections, possibly as a result of the widespread use of cephalosporins, which may select out resistant organ-isms such as enterococci in the fecal flora of the patients (12). Knowledge of the epidemiology of enterococci is limited by a lack of practical and simple typing methods for these bacteria. Traditional methods, such as bacteriocin typing (11), phage typing (11), serotyping (19), or biotyping (2), are only moder-ately useful (13, 16). In recent years, typing methods based on analysis of bacterial DNA have been successfully applied to enterococci (5, 13, 16). These methods, although highly dis-criminative and versatile, are rather laborious, especially when comparing data obtained from many isolates.
We have developed a simple, automated system for typing of bacterial isolates (the Phene Plate [PhP] system). The system is based on numerical analysis of biochemical reaction kinetics (biochemical fingerprinting) (7, 14). For fermenting bacteria, the system is highly discriminatory, is reproducible, and allows large numbers of isolates to be compared. We have previously used the PhP system for epidemiological and ecological studies of different groups of enterobacteria (6, 8, 14, 22), Aeromonas
spp. (10), and coagulase-negative staphylococci (21). In the present study, the PhP system was developed to type entero-coccal isolates. The PhP results were compared with those obtained by ribotyping and pulsed-field gel electrophoresis (PFGE) of DNA restriction fragments. Using the PhP system, we also compared enterococcal isolates from fecal samples of healthy individuals with isolates from the blood of neonates.
MATERIALS AND METHODS
Bacterial strains.In the present study, 243 isolates from the following three different collections of enterococci were included: (i) A collection of 170 unre-lated fecal isolates from healthy individuals from throughout Sweden (89 En-terococcus faecalis isolates, 27 EnEn-terococcus faecium isolates, and 54 other en-terococcal species), (ii) isolates of E. faecalis from the blood of Swedish neonates in 11 different hospitals (n528 isolates), and (iii) clinical isolates of E. faecalis which had previously been subject to DNA typing by PFGE (n545 isolates). Of the last group of 45 isolates, 31 isolates had been collected during periods of from 1 to 6 months from one hospital each in Thailand (collection IIIA) (18), Chile (collection IIIB) (16), Houston, Tex. (collection IIIC) (16), and Argentina (col-lection IIID) (15). The other 14 isolates wereb-lactamase-producing isolates collected during an 8-year period from various countries and various U.S. states (collection IIIE) (17). The five isolates from Argentina and one isolate from Houston were alsob-lactamase-producing isolates. A majority (36 of 45) of the isolates had previously been subjected to ribotyping (3).
Identification of the species of the isolates was done by using the criteria of Facklam and Collins (2) by the method of Gray et al. (4). Isolates that produced acid from lactose, mannitol, and sorbitol but not from arabinose or sorbose were identified as E. faecalis. Isolates which produced acid from arabinose, lactose, and mannitol but not from sorbitol or sorbose were identified as E. faecium.
Biochemical fingerprinting using the PhP system.The PhP system modified for typing of fecal streptococci (PhP-FS plate; BioSys inova, Stockholm, Sweden)
* Corresponding author. Mailing address: Laboratory for Bacteriol-ogy, Microbiology and Tumorbiology Center, Karolinska Institute, S-171 77 Stockholm, Sweden.
2812
on May 15, 2020 by guest
http://jcm.asm.org/
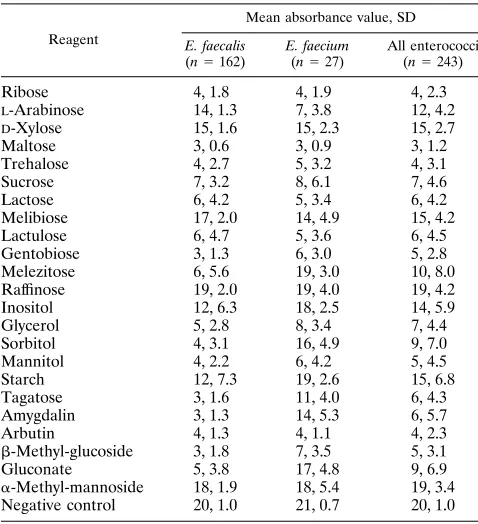
consists of 96-well microplates each with four sets of 24 dehydrated reagents (see Table 1). Twenty-three of the reagents had been selected from among a set of 96 reagents to yield the best discrimination and the best reproducibility in tests with 200 enterococcal isolates from various seawater and sewage water samples (data not shown). The 24th well contained a negative control.
The isolates to be tested were precultivated on nutrient agar plates. A 1-ml inoculation loop was used to suspend a small amount of bacteria into a medium containing 0.2% proteose peptone and 0.01% bromothymol blue, and 150ml of each bacterial suspension was dispensed into each of 24 wells of a PhP-FS plate. The inoculated plates were incubated at 378C for 3 days, and the A620of each well was measured after 16, 40, and 64 h of incubation by using a microplate reader. The absorbances were automatically transferred to a personal computer, multiplied by 10, and stored as integer values. After the last measurement, the mean value of the three readings was calculated for each reaction. The biochem-ical fingerprint of an isolate thus consisted of 24 numbers, each one ranging from 0 (acidic reaction) to 20 (neutral reaction).
The biochemical fingerprints of the isolates were compared pairwise, and the similarity between each pair was expressed as the correlation coefficient. This yielded a similarity matrix consisting of n3(n21)/2 correlation coefficients, where n is the number of isolates. The similarity matrix was clustered according to the unweighted-pair group method by using average linkages (UPGMA) to yield a dendrogram (20).
The reproducibility of the PhP-FS plate was evaluated by assaying 12 isolates in duplicate in the same assay (intra-assay reproducibility) and separately on two different occasions (interassay reproducibility). The similarity level defining iden-tity between isolates (the ID level) was defined as the mean correlation coeffi-cient between these duplicate assays of isolates minus 2 standard deviations of this mean (9). Isolates with a higher level of similarity than the ID level were assigned to the same PhP type. Isolates not identical to any other strains were named single PhP types.
All data processing, including optical readings and calculations of correlation coefficients as well as clustering and printing of dendrograms, was performed with PhP software (BioSys inova).
Ribotyping data.Ribotyping data for 36 of the 45 isolates from collection III were reported previously (3).
DNA typing by PFGE.Genomic DNA was prepared in agarose plugs as described previously (16). Slices of DNA were incubated overnight at 258C with SmaI (New England Biolabs, Beverly, Mass.). The agarose slice was then placed in the wells of 1.2% SeaPlaque GTG agarose gels (FMC, Rockland, Maine) in TBE buffer (0.089 M Tris-HCl, 0.089 M boric acid, 0.025 M EDTA), and electrophoresis was performed. Electrophoresis patterns were considered to be related, suggesting a common origin of the isolates, if their banding patterns differed by fewer than four bands. Some isolates were also analyzed with ApaI (New England Biolabs).
Comparison between typing methods.In order to calculate the diversity within a collection of strains, all isolates were compared pairwise. The number of pairwise comparisons yielding the same type was calculated, and that value was divided by the total number of comparisons. This yields the same result as that obtained from Simpson’s diversity index, which we used in our previous studies (9). The value of this index depends both on the number of different types identified and on the evenness of the distribution of isolates into different types. It is high (maximum value, 1.0) if most isolates belong to different types and is low (minimum value, 0) if one type dominates. The discriminatory power for the PhP typing systems was defined as the diversity among the unrelated isolates from collection I.
In order to be able to measure the agreement between PhP typing, ribotyping, and DNA typing for the isolates from collection III, we used the following calculation method. First, the diversity of the isolates was calculated by all three typing methods. Then, the isolates were compared pairwise by two typing meth-ods. Members of each pair of isolates which were either identical or different by both typing methods compared yielded a score of11 (which indicated good agreement). Members of each pair which were identical by the typing method showing the lowest diversity and different by the method showing the highest diversity also yielded a score of11 (good agreement). Finally, members of each pair which were different by the typing method showing the lowest diversity and identical by the method showing the highest diversity were given a score of21 (bad agreement). The sum of all scores was calculated and divided by the number of comparisons. By multiplying this value by 100, a value which is an estimate of the percent agreement between the two typing methods was obtained.
RESULTS
The PhP-FS system for typing of enterococci.Of the indi-vidual PhP-FS reagents selected, melezitose, sorbitol, amygda-lin, and gluconate yielded acidic reactions for most E. faecalis isolates but not for E. faecium isolates, andL-arabinose yielded acidic reactions for most E. faecium isolates but not for E.
faecalis isolates (Table 1). Thus, these five reagents best
dis-criminated between E. faecalis and E. faecium isolates. The intra-assay ID level was 0.975, and since all isolates studied
here were run in the same assay, this ID level was used for the data analysis. The interassay ID level was slightly lower (0.965) and should be used for defining the identities of isolates run in different assays. The diversity obtained for the 170 fecal iso-lates studied (collection I) was 0.97, and 107 different PhP types were identified (Table 2). E. faecium (collection Ib) and enterococci of other species (collection Ic) yielded greater diversity than E. faecalis (collection Ia, Table 2).
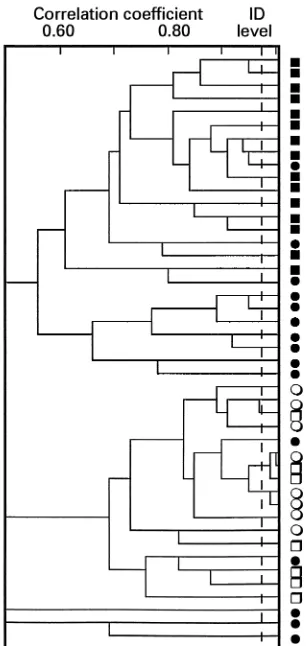
When clustering the data obtained by PhP typing of entero-cocci of various species, all E. faecalis isolates fell within one cluster, all E. faecium isolates fell into another, more hetero-geneous cluster (Fig. 1), and most of the unnamed enterococci did not belong to any of these clusters. Thus, in addition to being a typing system for enterococci, PhP typing might also be a valuable help for the identification of E. faecalis and E.
faecium species.
The stability of the biochemical fingerprints of enterococcal isolates was studied by subculturing 16 isolates on nutrient agar 20 times. The subcultured isolates and the parental isolates (stored at 48C) showed the same biochemical fingerprints. Thus, the PhP system showed a high degree of reproducibility and discriminatory power for enterococcal isolates, and the PhP types were stable over time.
Comparison of PhP typing, ribotyping, and DNA typing of
E. faecalis isolates. DNA typing by PFGE normally yielded
banding patterns showing 15 to 20 bands per strain, whereas ribotyping yielded only 7 to 10 bands (3). With the 45 E.
faecalis isolates from collection III, DNA typing discriminated
[image:2.612.315.554.82.344.2]27 types and PhP typing discriminated 19 types. Normally, when isolates were of the same DNA type, they were also of the same PhP type, and vice versa. An example of the relations between PhP types and DNA types is shown in Fig. 2. The two related DNA types, types C1 and C1a (from Chile), also showed two related PhP types which differed only in inositol fermentation (isolate CE39 was a fast inositol fermenter, whereas isolates CE13 and CE30 were negative for inositol
TABLE 1. Fermentation of PhP-FS reagents by enterococci
Reagent
Mean absorbance value, SD
E. faecalis (n5162)
E. faecium (n527)
All enterococci (n5243)
Ribose 4, 1.8 4, 1.9 4, 2.3
L-Arabinose 14, 1.3 7, 3.8 12, 4.2
D-Xylose 15, 1.6 15, 2.3 15, 2.7
Maltose 3, 0.6 3, 0.9 3, 1.2
Trehalose 4, 2.7 5, 3.2 4, 3.1
Sucrose 7, 3.2 8, 6.1 7, 4.6
Lactose 6, 4.2 5, 3.4 6, 4.2
Melibiose 17, 2.0 14, 4.9 15, 4.2
Lactulose 6, 4.7 5, 3.6 6, 4.5
Gentobiose 3, 1.3 6, 3.0 5, 2.8
Melezitose 6, 5.6 19, 3.0 10, 8.0
Raffinose 19, 2.0 19, 4.0 19, 4.2
Inositol 12, 6.3 18, 2.5 14, 5.9
Glycerol 5, 2.8 8, 3.4 7, 4.4
Sorbitol 4, 3.1 16, 4.9 9, 7.0
Mannitol 4, 2.2 6, 4.2 5, 4.5
Starch 12, 7.3 19, 2.6 15, 6.8
Tagatose 3, 1.6 11, 4.0 6, 4.3
Amygdalin 3, 1.3 14, 5.3 6, 5.7
Arbutin 4, 1.3 4, 1.1 4, 2.3
b-Methyl-glucoside 3, 1.8 7, 3.5 5, 3.1
Gluconate 5, 3.8 17, 4.8 9, 6.9
a-Methyl-mannoside 18, 1.9 18, 5.4 19, 3.4 Negative control 20, 1.0 21, 0.7 20, 1.0
on May 15, 2020 by guest
http://jcm.asm.org/
fermentation), whereas the three HH isolates (from Houston) showed DNA patterns and PhP types clearly different from those of the isolates from Chile. In some cases isolates with an identical PhP type had different DNA types, for example, iso-late CE36 in Fig. 2, which had the same PhP type as isoiso-late CE39 but a diffuse PFGE banding pattern which was not sim-ilar to that of CE39.
DNA typing and PhP typing combined identified 34 different types. Ribotyping of 36 of the isolates from collection III yielded only five different types and did not add to the number of types obtained by PhP-FS and DNA typing. The diversity was 0.94 for DNA typing, 0.92 for PhP typing, 0.56 for ribotyp-ing (Table 2), and 0.98 for DNA typribotyp-ing and PhP typribotyp-ing com-bined. The level of agreement between DNA typing and PhP typing was 96%, and that between ribotyping and PhP typing was 97%. Since DNA typing could discriminate isolates within ribotypes, but not vice versa, ribotyping had 100% agreement with DNA typing. In one case, ribotyping could discriminate between isolates of the same PhP type (isolates BE18 and BE81; Fig. 3) and showed 97% agreement with PhP typing. Thus, all three typing methods showed good agreement.
[image:3.612.57.556.84.218.2]The PhP types of E. faecalis isolates from one hospital each in Thailand, Chile, Texas, and Argentina (collections IIIA to IIID, respectively) were clustered to yield a separate dendro-gram for each collection, to which DNA types and ribotypes were added (Fig. 3). The majority of the isolates from Thailand belonged to a single PhP type (type A1) and the same ribotype (ribotype A). Four of the seven isolates belonging to PhP type A1 also had the same DNA type and ribotype. The isolates from Chile formed four PhP types, which, except for the DNA type of CE39, was in agreement with the types obtained by the other typing methods. Also, among the isolates from Texas, a high level of agreement between all three typing methods was found. The only exception was for isolate HH22, which be-longed to PhP type C1, but according to DNA type and ri-botype it was a single isolate. The five Argentinian isolates were of three closely related DNA types and also of three
FIG. 1. Dendrograms derived from UPGMA clustering of PhP types of some randomly selected enterococcal isolates from healthy individuals and from the blood of neonates with septicemia in Sweden.E, E. faecalis isolates from healthy individuals (collection Ia);h, E. faecalis isolates from blood of neonates with septicemia (collection II);■, E. faecium isolates from healthy individuals (col-lection Ib);F, other enterococcal species from healthy individuals (collection Ic).
[image:3.612.102.256.350.673.2]FIG. 2. Relations between PhP types and DNA types of seven E. faecalis isolates from Chile and Houston. The dendrogram was derived from UPGMA clustering of PhP data, and the electrophoretic pattern was obtained by PFGE of SmaI-digested chromosomal DNAs of the same isolates.
TABLE 2. Diversities of enterococci in different collections by different typing methods
Collection Origin Species No. of
isolates
Diversity (no. of types)
PhP-FS typing DNA typing Ribotyping
I Sweden, fecal Various 170 0.97 (107)
Ia Sweden, fecal E. faecalis 89 0.93 (32)
Ib Sweden, fecal E. faecium 27 0.99 (25)
Ic Sweden, fecal Other Enterococcus spp. 54 1.00 (50)
II Sweden, neonatal blood E. faecalis 28 0.93 (20)
III Worldwide E. faecalis 45a 0.92 (19) 0.94 (27) 0.56 (5)
IIIA Thailand E. faecalis 12 0.68 (6) 0.91 (9) 0.42 (3)
IIIB Chile E. faecalis 7 0.81 (4) 0.91 (6) 0.48 (2)
IIIC Houston E. faecalis 7 0.86 (5) 0.95 (6) 0.67 (3)
IIID Argentina E. faecalis 5 0.80 (3) 0.80 (3) 0.00 (1)
IIIE Worldwideb E. faecalis 20 0.93 (11) 0.64 (7) 0.53 (3)
a
Ribotyping was used to test 36 of the isolates. bb-Lactamase positive.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:3.612.318.553.584.692.2]closely related PhP types, but isolates with identical DNA types were slightly different according to their PhP types, and vice versa. The two PhP type D1 isolates were obtained in June and July, and the two PhP type D2 isolates were obtained in Oc-tober, whereas one isolate, belonging to a single PhP type, was obtained in April.
PhP typing of the 20b-lactamase-producing E. faecalis iso-lates yielded 11 types and a diversity of 0.94. Clustering of the PhP types of these isolates showed two distinct clusters, one containing the Argentinian isolates plus one U.S. isolate which was of the same ribotype as the Argentinian isolates and one cluster containing only U.S. isolates (Fig. 4). Two clusters and three single isolates were found by DNA typing. The two DNA clusters (named A1 and BCP) each contained several isolates which were similar to each other (differing by one or a few bands) and which also had similar or identical PhP types. The three isolates outside these DNA clusters also had single, unique PhP types.
Diversity of PhP types in the Swedish enterococcal collec-tions.PhP typing of the isolates from collection I (unrelated Swedish isolates belonging to various enterococcal species) yielded a diversity as high as 0.97 (Table 2). The E. faecalis isolates from collection I showed a lower diversity index (0.93), whereas E. faecium isolates and enterococci belonging to other species showed diversity indices of 0.99 or greater. These num-bers can thus be regarded as representative for PhP types
among unrelated enterococcal isolates. Collection II (E.
fae-calis isolates from the blood of neonates with septicemia in
Sweden) showed a diversity of 0.93, i.e., the same as that of unrelated E. faecalis isolates from healthy individuals, and the same PhP types which were dominant among the isolates from healthy individuals were also predominant among the neonatal blood isolates (Fig. 1). Thus, the invasive isolates seemed to make up a normal E. faecalis population, without the domi-nance of certain PhP types which could have been associated with virulence.
DISCUSSION
Traditional typing methods for bacteria based on phenotypic characters are often regarded as less discriminatory and as having lower levels of reproducibility than methods based on direct analysis of the bacterial genome. However, since the phenotype of a bacterial strain reflects its genetic contents, the performance of a phenotypic typing system may well match those of typing systems based on molecular techniques if stable and discriminatory phenotypic characteristics are measured. In the present study we have shown that our phenotyping method (the PhP-FS system) showed high degree of reproducibility and a high degree of discriminatory power for unrelated en-terococcal isolates and good agreement with DNA typing by PFGE and with ribotyping. The PhP types were also stable during subculturing.
Although biotyping is one of the traditional phenotypic methods for typing bacteria, it is not regarded as discrimina-tory enough for typing of enterococci. Most tests used for biotyping were originally developed for species identification. Like ribotyping, which measures conserved parts of the bacte-rial genome and thus may not be very discriminatory below the species level, the tests included in traditional biotyping often give similar results for all bacteria within a species. However, the biochemical versatility of many bacterial groups, including enterococci, is high, making the number of discriminatory re-actions to choose from rather extensive. Furthermore, tradi-tional biotyping schemes are based on qualitative measure-ments of the reactions (i.e., positive or negative), which may cause reproducibility problems because of various
interpreta-FIG. 3. Dendrograms derived from UPGMA clustering of the PhP types of E. faecalis isolates from hospitals in different countries.a
, single type (found only once in the collection).
FIG. 4. Dendrogram derived from UPGMA clustering of the PhP types of
b-lactamase-producing E. faecalis isolates from different countries and from different states in the United States.a
, single type (found for only one of the isolates;b
BCP,b-lactamase-producing E. faecalis common DNA type;c , NA, not assayed.
on May 15, 2020 by guest
http://jcm.asm.org/
tions of intermediate reactions, and fail to take advantage of the differences in kinetics and intensities of the reactions that may exist among strains of the same species. The PhP system for biochemical fingerprinting is based on quantitative mea-surements of the kinetics of biochemical reactions that have been selected for being highly discriminatory among isolates of the studied group of bacteria, and thus does not have the drawbacks of biotyping mentioned above.
A main advantage of PhP typing is that it is simple to per-form, and data processing and presentation are automated. Less than 8 h of labor, in total, including data interpretation and presentation, is required to assay 200 isolates, and prelim-inary results may be obtained after overnight incubation. Other advantages of PhP typing are that numerical data of high precision are generated, enabling cluster analysis and easy comparisons of large sets of old and new data. In contrast, molecular typing methods based on electrophoresis may have reproducibility problems when data generated on different oc-casions are compared. Thus, PhP typing is suitable as a screen-ing method, especially for large numbers of isolates, for which it can be used to select those isolates that need to be further typed by more cumbersome molecular typing techniques.
One important aspect of a typing system is how to define identity between isolates. For DNA typing this is usually done arbitrarily. For example, in the present study, enterococcal isolates that had identical PFGE patterns or that differed by one or a few bands were regarded as being of the same origin (3). For PhP typing, the ID level is defined statistically as the minimum similarity obtained from comparisons between inde-pendent duplicate assays. Isolates derived from the same an-cestor, but isolated far apart in time or geographically, may show some changes in their phenotypes and will thus be as-signed to different, but related, PhP types. This may be the case among the 20 b-lactamase-producing E. faecalis isolates. Among those isolates, several different, but similar, PhP types were found (Fig. 4), and the level of diversity among those isolates was as high as that among unrelated fecal isolates (diversity index, 0.93 for both collections). However, by arbi-trarily defining as related isolates with correlation coefficients to each other of .0.95, the diversity for the b -lactamase-producing isolates became low (diversity index, 0.77) as com-pared with that for unrelated E. faecalis isolates from fecal specimens (diversity index, 0.90) and isolates from the blood of infants (diversity index, 0.92). By using a similarity level of 0.95, the b-lactamase-producing isolates could be subdivided into two major, clearly distinct PhP clusters, one of which mainly contained isolates from Argentina and the other of which con-tained isolates only from the United States. Thus, our PhP data support the theory that theseb-lactamase-producing isolates consist of descendants of two different clones, one of which has spread to several hospitals in the United States (17). In con-trast, E. faecalis isolates causing neonatal septicemia in differ-ent Swedish hospitals seemed to represdiffer-ent a normal popula-tion of E. faecalis rather than descendants of certain virulent clones.
Overall, DNA typing discriminated 25% more types than PhP typing. E. faecalis isolates from different areas could not always be discriminated by PhP typing, but they could be dis-criminated by DNA typing. Similarly, isolates within the same DNA cluster could sometimes be of different PhP types, e.g., DNA type BCP. Isolates within the same collection that were slightly different from each other according to DNA type or PhP type were sometimes identical by the other method (see, for example, the Argentinian isolates; Fig. 3, collection IIID). Since the two typing methods measure strain variations in different ways, this outcome is quite normal. Those few isolates
that were identical by all three typing methods were usually from the same hospital and were isolated during the same period of time, thus indicating a direct transfer of bacteria between patients. We think that minor differences between isolates by either method represent variations within the same strain, which may be an important consideration, e.g., when following the spread of a certain strain in a hospital ward, but these differences are of less importance when isolates from different geographical areas or from different time periods are compared.
We conclude that the PhP-FS system is useful for typing enterococcal isolates, yielding results similar to those obtained by DNA typing with PFGE, and that it can be used alone or in combination with DNA typing for epidemiological and ecolog-ical studies of enterococci.
ACKNOWLEDGMENTS
We thank all Swedish laboratories which supplied enterococcal iso-lates.
This work was supported by the Swedish Medical Research Council, grant B93-16X-8302-06B, and by Karolinska Institute funds.
REFERENCES
1. Clausen, E. M., B. L. Green, and W. Litsky. 1977. Fecal streptococci: indi-cators of pollution, p. 247–264. In A. W. Hoadley and B. J. Dutka (ed), Bacterial indicators/health hazards associated with water. Standard STP 635. Americal Society for Testing and Materials, Philadelphia.
2. Facklam, R. R., and M. D. Collins. 1989. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 27:731–734.
3. Gordillo, M. E., K. V. Singh, and B. E. Murray. 1993. Comparisons of ribotyping and pulsed-field gel electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. J. Clin. Microbiol. 31:1570–1574. 4. Gray, J. W., D. Stewart, and J. S. Pedler. 1991. Species identification and
antibiotic susceptibility testing of enterococci isolated from hospitalized pa-tients. Antimicrob. Agents Chemother. 35:1943–1945.
5. Hall, L. M. C., B. Duke, M. Guiney, and R. Williams. 1992. Typing of Enterococcus species by DNA restriction fragment analysis. J. Clin. Micro-biol. 30:915–919.
6. Katouli, M., I. Ku¨hn, R. Wollin, and R. Mo¨llby.1992. Evaluation of the PhP system for biochemical-fingerprint typing of strains of Salmonella of serotype typhimurium. J. Med. Microbiol. 37:245–251.
7. Ku¨hn, I.1985. Biochemical fingerprinting of Escherichia coli: a simple method for epidemiological investigations. J. Microbiol. Methods 3:159– 170.
8. Ku¨hn, I., G. Allestam, T.-A. Stenstro¨m, and R. Mo¨llby.1991. Biochemical fingerprinting of water coliform bacteria, a new method for measuring the phenotypic diversity and for comparing different bacterial populations. Appl. Environ. Microbiol. 57:3171–3177.
9. Ku¨hn, I., A. Brauner, and R. Mo¨llby.1990. Evaluation of numerical typing systems for Escherichia coli using the API 50 CH and the PhP-EC systems as models. Epidemiol. Infect. 105:521–531.
10. Ku¨hn, I., T. Lindberg, K. Olsson, and T.-A. Stenstro¨m.1992. Biochemical fingerprinting for typing of Aeromonas strains from food and water. Lett. Appl. Bacteriol. 15:261–265.
11. Kuhnen, E., K. Rommelsheim, and L. Andries. 1987. Combined use of phage typing, enterococcinotyping and species differentiation as an effective epide-miological tool. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. Abt. 1 Orig. Reihe A 266:586–595.
12. Magnusson, C. R., and J. Cave. 1988. Nosocomial enterococcal infections: association with use of third generation cephalosporin antibiotics. Am. J. Infect. Control 16:241–245.
13. Miranda, A. G., K. V. Singh, and B. E. Murray. 1991. DNA fingerprinting of Enterococcus faecium by pulsed-field gel electrophoresis may be a useful epidemiologic tool. J. Clin. Microbiol. 29:2752–2757.
14. Mo¨llby, R., I. Ku¨hn, and M. Katouli.1993. Computerized biochemical fin-gerprinting—a new tool for typing of bacteria. Rev. Med. Microbiol. 4:231– 241.
15. Murray, B. E., H. A. Lopardo, E. A. Rubeglio, M. Frosolono, and K. V. Singh. 1992. Intrahospital spread of a single gentamicin-resistant,b -lactamase-producing strain of Enterococcus faecalis in Argentina. Antimicrob. Agents Chemother. 36:230–232.
16. Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Micro-biol. 28:2059–2063.
on May 15, 2020 by guest
http://jcm.asm.org/
17. Murray, B. E., K. V. Singh, S. M. Markowitz, H. A. Lopardo, J. E. Patterson,
M. J. Zervos, E. Rubeglio, G. M. Eliopolus, L. B. Rice, F. W. Goldstein, S. G. Jenkins, G. M. Caputo, R. Nasnas, L. S. Moore, E. S. Wong, and G. Wein-stock.1991. Evidence for clonal spread of a single strain ofb -lactamase-producing Enterococcus (Streptococus) faecalis to six hospitals in five states. J. Infect. Dis. 163:780–785.
18. Murray, B. E., J. Tsao, and J. Panida. 1983. Enterococci from Bangkok, Thailand, with high-level resistance to currently available aminoglycosides. Antimicrob. Agents Chemother. 23:799–802.
19. Sharpe, M. D., and P. M. F. Shattock. 1952. The serological typing of group
D streptococci associated with outbreaks of neonatal diarrhoea. J. Gen. Microbiol. 6:150–165.
20. Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. The W. H. Freeman Co., San Fransisco.
21. Thore, M., I. Ku¨hn, S. Lo¨fdahl, and L.-G. Burman.1990. Evaluation of four marker systems and epidemiology of drug resistant coagulase-negative skin staphylococci in an orthopaedic ward. Epidemiol. Infect. 105:95–105. 22. Tullus, K., B. Berglund, B. Fryklund, I. Ku¨hn, and L. G. Burman.1988.
Epidemiology of fecal strains of the family Enterobacteriaceae in 22 neonatal wards and influence of antibiotic policy. J. Clin. Microbiol. 26:1166–1170.