0095-1137/11/$12.00 doi:10.1128/JCM.00145-11
Copyright © 2011, American Society for Microbiology. All Rights Reserved.
Characterization of Occult Hepatitis B Virus Infection from
Blood Donors in China
䌤
Xin Zheng,
1† Xianlin Ye,
2† Ling Zhang,
1Wenjing Wang,
1Lifang Shuai,
3Anqi Wang,
1Jinfeng Zeng,
2Daniel Candotti,
4Jean-Pierre Allain,
1,5* and Chengyao Li
1*
Department of Transfusion Medicine, Southern Medical University, Guangzhou, China1; Shenzhen Blood Centre, Shenzhen,
China2; Guangzhou Military Centre of Disease Control, Guangzhou, China3; National Health Service Blood and
Transplant, Cambridge Blood Centre, Cambridge, England4; and Department of Hematology,
University of Cambridge, Cambridge, United Kingdom5
Received 24 January 2011/Returned for modification 7 March 2011/Accepted 10 March 2011
Prevalence and characteristics of occult hepatitis B virus (HBV) infection (OBI) of genotypes B and C prevalent in China have not been extensively explored. Characterization of OBI strains obtained from Chinese blood donors was based on clinical and serological analyses, follow-up testing, and sequence analyses. Twenty-eight samples from 165,371 HBV surface antigen (HBsAg)-negative plasmas were confirmed HBsAg negative and DNA positive(HBsAgⴚ/DNAⴙ), of which 22 were classified as OBIs and 6 as window period infections. The OBI incidence was 1:7,517 in blood donors, whose ages ranged between 20 and 45 years (median, 28 years). OBI donors had normal alanine aminotransferase (ALT) levels and low viral loads ranging between unquantifiable amounts and 178 IU/ml (median, 14 IU/ml). Sequences from 21 basic core promoter/precore (BCP/PC) regions, five whole genomes, and two additional pre-S/S regions from OBI strains were compared to genotypes B and C HBsAgⴙreference strains. Eighty-six percent (6/7) of OBI strains were genotype C. Deletions, insertions, stop codons, and substitutions were detected in 15/21 (71%) core regulatory elements of OBI strains. Critical mutations were found in the core proteins of 5/5 OBI strains in parallel with random substitutions in pre-S/S proteins from 6/7 (86%) OBI strains. Critical mutations in core regulatory elements and core proteins might affect OBI genotype B and C strain replication. That there were few S protein substitutions suggests a minor role of the host immune defenses in OBI occurrence.
Hepatitis B virus (HBV) infection is a major threat to hu-man health worldwide, especially to people living in devel-oping countries. The number of HBV carriers with detect-able surface antigen (HBsAg) is estimated to be over 300 million globally, while there are around 100 million HBV-infected individuals in mainland China (12). HBV serolog-ical testing is widely used in controlling the risk of HBV transmission among people who donate blood or have health examinations in China. However, some studies re-cently showed HBV DNA at low levels detected in HBsAg-negative individuals, some of whom transmitted HBV infec-tions to recipients of blood transfusions (10). Occult hepatitis B virus infection (OBI) is defined as the detection of HBV DNA in the serum or liver tissue of patients who test negative for HBsAg (15). Currently, testing for HBsAg is performed for screening of HBV infection in blood do-nors in China, but nucleic acid testing (NAT) is not used.
A characterization of OBI strains of genotypes A1, A2, D, and E in comparison with wild-type HBV in samples obtained from European and African blood donors was recently pub-lished. The mechanisms for generating OBI strains were
mainly related to either imperfect immune control or to genetic viral defects and appeared largely genotype dependent (1, 4). However, analysis of OBI strains of genotypes B and C prevalent in Southeast Asia and China has not been extensively explored, particularly in terms of molecular characterization (14, 20).
This study provides new information regarding the preva-lence and the molecular analysis of OBI strains of genotypes B and C from samples obtained from asymptomatic blood donors tested in Shenzhen, China.
MATERIALS AND METHODS
Blood center and sample screening.Donor blood samples were collected by the Shenzhen blood center between August 2003 and December 2009. Donor plasmas were individually screened twice for HBsAg, anti-hepatitis C virus (HCV), and anti-HIV antibodies using domestic and international commercial enzyme immunoassays (EIA), including Xiamen Xinchuang HBsAg EIA (Xia-men Xinchuang Scientific Ltd., Xia(Xia-men, China), Abbott Auszyme HBsAg (Abbott Laboratories, Abbott Park, IL), Sino-America Huamei anti-HCV EIA (Sino-America Huamei Biotechnology Ltd., Luoyang, China), Ortho anti-HCV EIA (Ortho-Clinical Diagnostics, Inc., Raritan, NJ), Zhuhai Lizhu anti-HIV EIA (Zhuhai Lizhu Biotechnology Ltd., Zhuhai, China), and Melia anti-HIV-1 and -2 EIA (Organon Teknika, Boxtel, Netherlands). The level of alanine aminotrans-ferase (ALT) was also measured for each blood sample with a ReflotronPlus assay (Roche Diagnostics, Mannheim, Germany).
Nucleic acid testing. Donor plasmas negative for HBsAg, anti-HCV, and anti-HIV were tested individually for nucleic acids of HBV, HCV, and HIV-1 with a Roche Cobas Ampliscreen assay (Roche), Kehua Real-Time PCR assay (Kehua Biotechnology Ltd., Shanghai, China), or Procleix Ultrio assay (Novartis, Emeryville, CA).
Serological testing.HBsAg reactivity was confirmed with a highly sensitive chemiluminescent microplate immunoassay ([CMIA] Architect HBsAg assay; Abbott laboratories) used according to the manufacturer’s instructions. Anti-HBs, hepatitis B e antigen (HBeAg), anti-HBe, and anti-HBV core protein * Corresponding author. Mailing address for Chengyao Li:
Depart-ment of Transfusion Medicine, Southern Medical University, Guang-zhou, China. Phone and fax: 86 20 61648466. E-mail: chengyaoli @hotmail.com. Mailing address for Jean-Pierre Allain: Department of Hematology, University of Cambridge, Cambridge Blood Centre, Long Road, Cambridge CB2 2PT, United Kingdom. Phone: 44 1223 588044. Fax: 44 1223 588155. E-mail: jpa1000@cam.ac.uk.
† X.Z. and Z.Y. contributed equally to this work.
䌤Published ahead of print on 16 March 2011.
1730
on May 16, 2020 by guest
http://jcm.asm.org/
(HBc) were detected with Kehua HBV EIAs (Kehua). Anti-HBs was quantified in IU/liter with an anti-HBs electrochemiluminescence immunoassay (Roche).
HBV DNA quantification, amplification, and sequencing.HBV DNA load was determined by quantitative PCR (qPCR) as described previously (4, 21). Viral DNA was extracted from 0.4 to 1.0 ml of plasma (HighPure; Roche). The HBV basic core promoter/precore region (BCP/PC; nucleotide [nt] 1679 to 1973), the whole genome minus 53 bp (nt 1804 to 1856) in the precore region (3,162 bp), and the pre-S/S region (nt 2802 to 1020) were amplified using nested PCRs sequentially to obtain either the whole genome or fragments of the HBV genome (5, 21). Whole-genome amplicons were cloned in pMD20-T vector (TaKaRa Biotechnology Ltd., Dalian, China), and fragments of pre-S/S were cloned in the original TA cloning kit. Amplified BCP/PC products and two to six clones were sent for sequencing to Shanghai Invitrogen Co., Ltd. (Guangzhou, China).
The second WHO international standard for HBV DNA (National Institute for Biological Standards and Control [NIBSC], Potters Bar, United Kingdom) containing 5⫻105
IU per vial (NIBSC code 97/750) was kindly provided by Qingtao Song (Xiamen Xinchuang) for quantification of HBV DNA.
Occult HBV DNA analyses.Alignment and phylogenetic analyses were per-formed on a consensus sequence from each OBI plasma sample using the neighbor-joining (NJ) method and pre-S/S (1,201 bp) sequences (4). Control HBV wild-type sequences were mostly Chinese strains obtained from HBsAg-positive (HBsAg⫹) strains selected from the GenBank database. Accession numbers for genotype B strains were AB205119, AB287329, AP011084, AY167089, D00300, DQ448620, DQ448625, DQ993703, DQ993709, EU305548, EU306701, EU306705, EU306706, FJ386582, FJ386642 and FJ787444; for genotype C strains the accession numbers were AP011098, DQ089793, DQ478899, DQ478900, DQ993691, DQ993692, DQ993693, EU306720, EU306721, EU916238, EU916240, EU939540, EU939652, FJ386575, FJ386576, FJ386627, FJ386685, FJ562218, FJ562232, FJ562244, X04615, X75656, and X75665.
Diversity was defined as the mean value for a pairwise difference of sequence between OBI and HBV wild-type consensus sequences within the same group, calculated as the number of nucleotide or amino acid differences between two individual sequences (4).
Statistical analyses.SPSS software (version 13.0) was used for statistical anal-yses. Categorical variables were compared by using Fisher’s exact test and, for continuous variables, a nonparametric Mann-Whitney test.
RESULTS
Sample classification and characterization. A total of 165,371 donor plasma samples from Shenzhen Blood Center were screened as HBsAg negative (HBsAg⫺) with one inter-national and one domestic commercial EIA kit. Thirty-three plasmas were detected as HBV DNA positive (DNA⫹) by individual-sample NAT. Twenty-eight of 33 plasmas were con-firmed as HBsAg⫺ and DNA⫹ (HBsAg⫺/DNA⫹) by the CMIA for HBsAg and nested PCR for BCP/PC. Twenty of 33 blood donors with an index sample determined to be HBsAg⫺/ DNA⫹were recalled for follow-up testing of HBV infection markers 2 to 221 days after index donation.
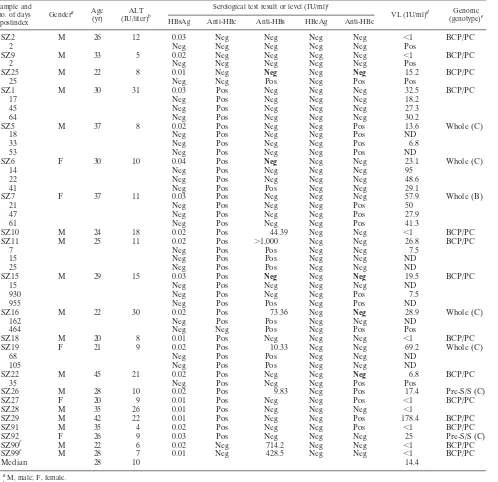
A total of 22/28 index samples determined to be HBsAg⫺/ DNA⫹were classified as OBIs (Table 1). Seromarker-negative SZ2 and SZ9 donors tested positive for HBV DNA by a Roche PCR-enzyme-linked immunosorbent assay (ELISA) and nested PCR in follow-up samples and were classified as seronegative or primary OBIs (1, 15). Donor SZ25 with negative seromarkers was found reactive for both anti-HBs and anti-HBe 25 days after the index sample was obtained (postindex), and this sample was clas-sified as OBI. The other 19 samples were confirmed as OBIs by serological markers or follow-up testing. Donors SZ90 and SZ99 were high-level anti-HBs⫹only (⬎428 IU/liter) and had been vaccinated for HBV 6 months and 3 years prior to blood dona-tion, respectively.
Four index samples (SZ14, SZ17, SZ23, and SZ200) with no serological markers and a viral load of⬎200 IU/ml were con-firmed HBV window period (WP) infections following
sero-conversion to HBsAg or HBeAg (1st WP) (Table 2). Two samples (SZ4 and SZ12) carrying anti-HBe⫹and anti-HBc⫹ seroconverted to HBsAg and were classified as secondary win-dow period infections (2nd WP). Sample SZ3 that was both anti-HBe⫹and anti-HBc⫹was initially classified as HBsAg⫺ by two EIAs but was weakly reactive (0.13 IU/ml) by CMIA and was reclassified as chronic infection. Sample SZ24 was found to be HBsAg⫺but HBeAg⫹and was considered chronic infection. Samples SZ13, SZ20, and SZ21 carrying either anti-HBs⫹ or anti-HBc⫹ but without confirmed HBV DNA by BCP/PC nested PCR were considered false positives.
The prevalence of confirmed OBI was 1:7,517 (22/165,371) in Shenzhen blood donors (Table 1). All 22 participants were between 20 and 45 years old (median, 28 years) and had nor-mal ALT levels (⬍40 IU/liter). Seventeen samples (77.3%) were anti-HBc⫹, seven (31.8%) were anti-HBs⫹(five with anti-HBc), five (22.7%) were anti-HBe⫹(all with anti-HBc), and three (13.6%) were negative for all seromarkers at initial test-ing. Viral load of OBI strains ranged between unquantifiable (defined as⬍1 IU/ml) and 178.4 IU/ml (median, 14.4 IU/ml). There was no significant difference in viral load between anti-HBs- or anti-HBe-positive and -negative OBI strains (P ⫽
0.798 orP⫽0.903, respectively).
Phylogenetic analysis of five full-length genome (SZ5, SZ6, SZ7, SZ16, and SZ19) and two additional pre-S/S (SZ26 and SZ92) sequences identified one (14.3%) genotype B (OBIB)
and six (85.7%) genotype C (OBIC) OBI strains (Table 1).
Analysis of regulatory elements of occult HBV sequences.
The consensus nucleotide sequences of regulatory elements from OBIB and OBIC strains were compared to consensus
sequences from 16 genotype B or 23 genotype C HBsAg⫹ strains, respectively. Promoters (for pre-S1, pre-S2/S, core, and X proteins), regulatory sequences (core upstream regulatory sequence [CURS] and direct repeat sequences [DR]), enhanc-ers (ENH), and negative regulatory elements (NRE) were analyzed. DR1, DR2, and NRE were conserved. Significantly higher mean diversity was observed in the S promoter region (SP) and ENH regulatory elements from the OBIC sequence
(Pⱕ0.004 to 0.0001) but not in CURS, BCP, and X promoter region (XP) sequences between OBIC and HBsAg⫹ strains
(P⫽0.025 to 0.846) (Table 3).
Mutations were observed in CURS and BCP regions in 71.4% (15/21) of OBI sequences but less frequently in HBsAg⫹control sequences (Table 4). Thirteen of 21 (61.9%) OBI sequences pre-sented CURS (nt 1643 to 1741) mutations consisting of a dele-tion, inserdele-tion, or single or multiple substitutions. OBI samples SZ22 and SZ92 had one nucleotide deletion at the same site, nt 1689; OBI SZ9 had an insertion of C at nt 1716. A total of 11 mutations were detected in the CURS region of 11 OBI strains and were present in several strains at nt 1727, 1730, 1703, 1719, and 1726. In the BCP region, mutations were detected at six sites in 8/21 (38.1%) OBI strains, including G1742A (SZ5 and SZ15), A1752G (SZ5, SZ15, SZ16, and SZ18), A1762T and G1764A (SZ6), G1799C or C1799G (SZ5, SZ16, SZ19, and SZ92), and A1846T (SZ91).
In the epsilon domain eight OBI strains had mutations, but only T1893A (SZ99) had a possible structural impact on the bulge formation.
Analyses of occult HBV core, X, envelope, and polymerase proteins.Seven OBI amino acid sequences were aligned with
on May 16, 2020 by guest
http://jcm.asm.org/
the genotype B or genotype C HBsAg⫹control strains. The mean number of amino acid substitutions in pre-C/C, pre-S/S, and polymerase (Pol) regions from the OBIC sequence was higher than in wild-type strains (Pⱕ0.013 to 0.0001) (Table 3). In the precore region of five OBI strains, SZ7 had a V17F mutation, and SZ16 had a stop codon at amino acid (aa) 28.
[image:3.585.46.544.81.563.2]Critical mutations in the core region of all five OBI strains associated with B cell, T helper (Th) cell, and cytotoxic T lymphocyte (CTL) epitopes might theoretically influence im-munological HBV epitope recognition (Fig. 1) (3, 16). The common mutations I97(L/T) (SZ7, SZ6, and SZ19), P130(T/S) (SZ6 and SZ16), S87G (SZ7 and SZ6), and P135(S/Q) (SZ7
TABLE 1. Characterization of OBI strains from China Shenzhen blood donors
Sample and no. of days postindex
Gendera Age
(yr)
ALT (IU/liter)b
Serological test result or level (IU/ml)c
VL (IU/ml)d Genome
(genotype)e
HBsAg Anti-HBc Anti-HBs HBeAg Anti-HBe
SZ2 M 26 12 0.03 Neg Neg Neg Neg ⬍1 BCP/PC
2 Neg Neg Neg Neg Neg Pos
SZ9 M 33 5 0.02 Neg Neg Neg Neg ⬍1 BCP/PC
2 Neg Neg Neg Neg Neg Pos
SZ25 M 22 8 0.01 Neg Neg Neg Neg 15.2 BCP/PC
25 Neg Neg Pos Neg Pos Pos
SZ1 M 30 31 0.03 Pos Neg Neg Neg 32.5 BCP/PC
17 Neg Pos Neg Neg Neg 18.2
45 Neg Pos Neg Neg Neg 27.3
64 Neg Pos Neg Neg Neg 30.2
SZ5 M 37 8 0.02 Pos Neg Neg Pos 13.6 Whole (C)
18 Neg Pos Neg Neg Pos ND
33 Neg Pos Neg Neg Pos 6.8
53 Neg Pos Neg Neg Pos ND
SZ6 F 30 10 0.04 Pos Neg Neg Neg 23.1 Whole (C)
14 Neg Pos Neg Neg Neg 95
22 Neg Pos Neg Neg Neg 48.6
41 Neg Pos Pos Neg Neg 29.1
SZ7 F 37 11 0.03 Pos Neg Neg Neg 57.9 Whole (B)
21 Neg Pos Neg Neg Pos 50
47 Neg Pos Neg Neg Pos 27.9
61 Neg Pos Neg Neg Pos 41.3
SZ10 M 24 18 0.02 Pos 44.39 Neg Neg ⬍1 BCP/PC
SZ11 M 25 11 0.02 Pos ⬎1,000 Neg Neg 26.8 BCP/PC
7 Neg Pos Pos Neg Neg 7.5
15 Neg Pos Pos Neg Neg ND
25 Neg Pos Pos Neg Neg ND
SZ15 M 29 15 0.03 Pos Neg Neg Neg 19.5 BCP/PC
15 Neg Pos Neg Neg Neg ND
930 Neg Pos Neg Neg Pos 7.5
955 Neg Pos Pos Neg Pos ND
SZ16 M 22 30 0.02 Pos 73.36 Neg Neg 28.9 Whole (C)
162 Neg Pos Pos Neg Neg ND
464 Neg Neg Pos Neg Pos Pos
SZ18 M 20 8 0.01 Pos Neg Neg Neg ⬍1 BCP/PC
SZ19 F 21 9 0.02 Pos 10.33 Neg Neg 69.2 Whole (C)
68 Neg Pos Pos Neg Neg ND
105 Neg Pos Pos Neg Neg ND
SZ22 M 45 21 0.02 Pos Neg Neg Neg 6.8 BCP/PC
35 Neg Pos Neg Neg Pos Pos
SZ26 M 28 10 0.02 Pos 9.83 Neg Pos 17.4 Pre-S/S (C)
SZ27 F 20 9 0.01 Pos Neg Neg Pos ⬍1 BCP/PC
SZ28 M 35 26 0.01 Pos Neg Neg Neg ⬍1
SZ29 M 42 22 0.01 Pos Neg Neg Pos 178.4 BCP/PC
SZ91 M 35 4 0.02 Pos Neg Neg Pos ⬍1 BCP/PC
SZ92 F 26 9 0.03 Pos Neg Neg Neg 25 Pre-S/S (C)
SZ90f M 22 6 0.02 Neg 714.2 Neg Neg ⬍1 BCP/PC
SZ99f M 28 7 0.01 Neg 428.5 Neg Neg ⬍1 BCP/PC
Median 28 10 14.4
aM, male; F, female.
bALT level was tested by ReflotronPlus (Roche) in Nanfang hospital, Guangzhou, China.
cSerological markers of HBV except HBsAg were detected initially by Kehua HBV EIAs and retested by Architect HBsAg in Nanfang hospital. An HBsAg level
of⬍0.05 IU/ml was considered negative. Anti-HBs⫹was quantified in IU/liter by the Cobas anti-HBs electro-chemiluminescence immunoassay (Roche) in Nanfang hospital. Results in boldface indicate seroconversion during follow-up. Neg, negative; Pos, positive.
dViral load was quantified by qPCR against an international HBV standard. A value of⬍1 IU/ml indicates a sample with no threshold cycle value but confirmed
by nested PCR (Pos). ND, not done.
eOBI genomes were amplified by nested PCRs and sequenced by Invitrogen. The whole genome or fragments of HBV DNA were amplified by the nested PCRs,
and genotyping was phylogenetically performed from pre-S/S sequences aligned with references. ND, not done.
fDonors of index sample SZ90 and SZ99 were vaccinated 6 and 36 months prior to indexing, respectively.
on May 16, 2020 by guest
http://jcm.asm.org/
and SZ5) were found in 5/5 (100%) core regions of OBI strains. A specific substitution of L16I, S21L, S26A, P50H, L55I, L60V, M66V, L95I, or T142M was present in the core regions of all sequenced OBI strains but not of WP or chronic infection wild-type HBV strains.
In the X region, a mutation of A1762T in BCP led to a substitution of K130M in SZ6. Mutation V88(A/N) was de-tected in SZ6 and SZ19, and V5L and Q87G were dede-tected in SZ19. One nucleotide deletion at nt 1689 and insertion at nt 1716 in the CURS regions caused an X protein open reading frame (ORF) shift and aberrant translation in OBI samples SZ22, SZ92, and SZ9.
Mutations in the pre-S/S region of OBI strains were exam-ined with regard to the known epitopes of CTL, Th, and B cells and the major hydrophilic region (MHR) and “a” determinant (Fig. 2) (4, 7, 14). OBI MHR and “a” regions were relatively conserved, and no hot spot mutations were found. Few muta-tions occurred at sites of subtype conversion of adw and adr
corresponding to genotypes B and C. Substitutions I110L and I126T were found in samples SZ7 (OBIB) and SZ5 and SZ6
(OBIC), and A159V in SZ7 was presumably related to subtype (Fig. 2D). SZ5 presented substitutions T118M and W165R in MHR. Frequent substitutions were observed in OBI strain S protein at aa 2 to 5 in 4/7 (57%) (SZ7, SZ16, SZ19, and SZ26), in the middle position at aa 88 to 104 within CTL epitope clusters 9 and 10 in 4/7 (57%) OBIs (Fig. 2C), and at C-ter-minal aa 210 to 225 within the CTL epitope clusters 13 and 14 in 6/7 (85.7%) OBI strains (Fig. 2D). Similarly, frequent sub-stitutions were found in pre-S2 protein at the initial position of aa 5 to 9 within CTL epitope cluster 2 overlapping B-cell epitope cluster 8 from 5/7 (71.4%) OBI strains (Fig. 2B). SZ7 and SZ5 had a pre-S2 mutated start codon. SZ92 had two mutations (pre-sI84V and pre-sA90V) in pre-S1 that were also found in SZ7, SZ5, SZ6, and SZ16 (Fig. 1A).
[image:4.585.44.545.81.439.2]The mass random substitutions in the polymerase regions of OBIC were observed at a frequency ranging from 11 to 47
TABLE 2. Classification of HBV DNA yield from non-OBI index samples
Infection classification, sample,
and no. of days postindex
Gendera Age
(yr)
ALT (IU/liter)b
Serological test result or level (IU/ml)c
VL
(IU/ml)d Genome
(genotype)e
HBsAg Anti-HBc Anti-HBs HBeAg Anti-HBe
1st WP
SZ14 F 24 24 0.02 Neg Neg Neg Neg 224.6 BCP/PC
12 Neg Neg Neg Neg Neg 262
38 Pos Neg Neg Neg Neg 121
47 Neg Pos Pos Neg Neg ND
SZ17 M 26 18 0.02 Neg Neg Neg Neg 925.5 Whole (C)
45 Neg Neg Neg Pos Pos 23.2
SZ23 M 19 13 0.03 Neg Neg Neg Neg 675 Whole (B)
13 Pos Neg Neg Neg Neg 23
20 Pos Pos Neg Neg Neg 2429
127 Neg Neg Neg Pos Pos 85
SZ200 F 41 10 0.04 Neg Neg Neg Neg 283.8 Whole (B)
5 Neg Neg Pos Pos Pos 732
2nd WP or chronic
SZ4 M 29 23 0.02 Pos Neg Neg Pos 51.8 BCP/PC
21 Neg Pos Neg Neg Pos ND
45 Pos Pos Neg Neg Pos ND
91 Pos Pos Neg Neg Pos ND
156 pos Pos Neg Neg Pos ND
SZ12 M 25 22 0.03 Pos Neg Neg Pos ⬍1
8 Pos Pos Neg Neg Pos Pos
SZ3 M 42 11 0.13 Pos Neg Neg Pos 266.1 Whole (B)
38 Pos Pos Neg Neg Pos 323.2
60 Neg Pos Pos Neg Pos ND
121 Neg Pos Pos Neg Pos ND
166 Neg Pos Pos Neg Pos ND
221 Neg Pos Pos Neg Pos ND
SZ24 F 19 11 0.01 Pos 27.61 Pos Neg 7,321 Whole (C)
False positive
SZ13 M 34 20 0.02 Pos Neg Neg Neg Neg
SZ20 F 21 18 0.03 Neg Pos Neg Neg Neg
SZ21 F 28 25 0.02 Neg Pos Neg Neg Neg
a
M, male; F, female.
b
ALT level was tested by ReflotronPlus (Roche) in Nanfang hospital, Guangzhou, China.
c
Serological markers of HBV except HBsAg were detected initially by Kehua HBV EIAs and retested by Architect HBsAg in Nanfang hospital. An HBsAg level of⬍0.05 IU/ml was considered negative. Anti-HBs⫹was quantified in IU/liter by the Cobas anti-HBs electro-chemiluminescence immunoassay (Roche) in Nanfang hospital. Results in boldface indicate seroconversion during follow-up. Neg, negative; Pos, positive.
d
Viral load was quantified by qPCR against an international HBV standard. A value of⬍1 IU/ml indicates a sample with no threshold cycle value but confirmed by nested PCR (Pos). ND, not done.
e
OBI genomes were amplified by nested PCRs and sequenced by Invitrogen. The whole genome or fragments of HBV DNA were amplified by the nested PCRs, and genotyping was phylogenetically performed from pre-S/S sequences aligned with references.
on May 16, 2020 by guest
http://jcm.asm.org/
(Table 3), and no well-documented hot spot mutations were found in YMDD and FLLA motifs.
DISCUSSION
The prevalence of HBsAg in the Chinese population has declined to 7.2% due to nationwide HBV vaccination, but approximately 100 million HBV chronically infected
individu-als live in China (12). The new issue of occult HBV infection uncovered by NAT has become a concern in transfusion and in patients although little is known of OBI in China, where ge-notype B and gege-notype C are dominant (12, 20).
[image:5.585.43.542.82.206.2]The epidemiology of HBV infection in the Shenzhen blood donor population is somewhat reflective of that in China as a whole, except for a lower prevalence of HBsAg (12). The prevalence of OBI cases in Shenzhen was therefore expected
TABLE 3. Intra- and intergroup diversity analyses between OBICand HBsAg⫹sequencesa
Regulatory region sequence diversity Protein sequence diversity
Genome
regionb OBIC(n⫽4)
HBsAg⫹
(n⫽23) Pvalue
Genome
regionc OBIC(n⫽4)
HBsAg⫹
(n⫽23) Pvalue
ENH1 6.0 (4–9) 2.8 (0–10) 0.004 Pre-S/S 11.7 (2–22)d 3.6 (0–14) ⬍0.0001
SP1 14.5 (4–23) 6.2 (1–19) ⬍0.0001 Pre-S1 4.3 (2–7) 0.7 (0–3) ⬍0.0001
SP2 9.0 (5–17) 2.7 (0–9) ⬍0.0001 Pre-S2 0.7 (0–1) 0.9 (0–7) 0.630
S 7.0 (0–14) 1.9 (0–6) ⬍0.0001
ENH2 6.0 (4–10) 2.7 (0–7) 0.001 MHR 0.7 (0–3) 0.4 (0–1) 0.322
CURS 3.0 (2–4) 1.3 (0–4) 0.025 Pre-C/C 3.5 (2–5) 1.5 (0–5) 0.013
BCP 1.5 (0–3)d 1.4 (0–4) 0.846 Pol 23.3 (11–47) 8.9 (2–35) ⬍0.0001
Pol (RT) 4.5 (2–7) 1.5 (0–5) 0.001
XP 2.5 (1–3) 2.2 (0–5) 0.713 X 8.0 (5–15) 3.0 (0–7) ⬍0.0001
a
Regions with regulatory functions in HBV are presented separately. Average intragroup diversity was calculated as the number of nucleotide (regulatory region sequences) or amino acid (protein sequences) substitution differences between OBI and reference sequences. The range for frequency of nucleotide or amino acid substitutions is in parentheses.n, number of complete sequences from OBICor references used in analysis (except as noted). The difference of mean diversity between
OBICand reference sequences was calculated as thePvalue by Fisher’s Exact test.
b
SP1 (nt 2219 to 2780) and SP2 (nt 2809 to 3152), promoters for pre-S1 and pre-S2/S; XP (nt 1239 to 1376), promoter for X region; CURS (nt 1643 to 1741), core upstream regulatory sequence; BCP (nt 1742 to 1849), basal core promoter; ENH2 (nt 1627 to 1774), enhancer II; ENH1 (nt 1071 to 1238), enhancer I.
c
MHR, major hydrophilic region of S protein; Pol, polymerase; Pol (RT), reverse transcriptase; X, HBx.
d n⫽6.
TABLE 4. Site mutations in core promoter of OBI strains
Sample and or genotype
CURS (nt 1643–1741) sequence at position:a
BCP (nt 1742–1849) sequence at position:a
1652 1673 1679 1689 1703 1708 1716 1718 1719 1725 1726 1727 1730 1742 1752 1762 1764 1799 1846
HBsAg⫹ B/Cb
G/A T/C A A A T T T G A C/A T/A G/C G A A G G/C A
SZ2 NA NA G T G
SZ5 (C) /T /G A G /G
SZ6 (C) G T /G T A
SZ7 (B) A/
SZ9 NA NA Cb
SZ10 NA NA
SZ11 NA NA C
SZ15 NA NA A G
SZ16 (C) /C /T /G G /G
SZ18 NA NA G
SZ19 (C) /G /T /T /G /G
SZ22 NA NA Xc
SZ25 NA NA
SZ26 (C) NA NA T
SZ27 NA NA
SZ28 NA NA A
SZ29 NA NA
SZ90 NA NA
SZ91 NA NA G T
SZ92 (C) NA NA Xc /T /G /G
SZ99 NA NA
a
Positions of mutation sites within CURS and BCP are presented according to nucleotide numbering from EcoRI. NA, not available for nt 1643 to 1678 from BCP/PC amplicons of 16 OBI strains. HBsAg⫹B/C represents the nucleotide consensus sequence (in boldface) of genotype B or C (B/C) reference strains; their average diversity rates are 0.82% and 1.4%, respectively. The site mutations of OBI strains are indicated at genotype B/C positions of the references. The consensus at nt 1752 was 95.7% (22/23) nucleotide A within genotype C and 43.8% (7/16) A or 56.2% (9/16) G within genotype B.
b
C insertion in OBI SZ9.
c
X, deletion.
on May 16, 2020 by guest
http://jcm.asm.org/
[image:5.585.43.538.432.676.2]to be lower than average in China, but the frequency of 1:7,517 was similar to frequencies reported (median 1:9,819 and 1:8,209) in European or South African blood donors (1, 4). This is surprising considering a lower prevalence in Europe and a similar level in South Africa. In Hong Kong, close geo-graphically to Shenzhen, 80 OBI cases were identified in 357,440 blood donations, or 1:4,468, using the Novartis Ultrio screening method for individual donations. This apparently higher frequency might be related to a higher sensitivity of the screening method (6).
The ratio of male to female donors infected with OBI was 3.4:1 (17:5). The median age of 28 years of donors with OBI reflected the blood donor population in Shenzhen but was considerably lower than that of European blood donors with OBI of genotype A2 or D and of South African donors with OBI of genotype A1 (1, 4). As elsewhere, donors with OBI had normal ALT levels and viral loads below 200 IU/ml (median, 14.4 IU/ml) and carried anti-HBc⫹(77.3%) with anti-HBs or anti-HBs⫹only (9%). Compared to European data, the asso-ciation of anti-HBc and anti-HBs appears less frequent in Shenzhen’s donors but primary OBI is more frequent (4).
The availability of follow-up samples from 20/30 samples was of considerable help in ensuring the classification of samples and informing the donors, separating OBIs from window pe-riod and doubtful chronic infections, and, within OBIs, distin-guishing primary OBIs from other types (Tables 1 and 2). Among the latter, follow-up data showed the relative stability of HBV DNA detectability and load (Table 1).
Genomic amplification was relatively unsuccessful (12/30 for pre-S/S) in the context of low, often not quantifiable, viral load. Therefore, the genotype was determined in only 27% of OBI strains.
Mutations in core regulatory elements involved in viral replication were described previously (8, 13). Compared to HBsAg⫹strains, deletions, insertions, and substitutions at sin-gle or multiple sites were observed in the CURS and BCP (15/21) regions of OBI strains (Table 4). Mutations A1752G and A1762T (five OBI strains) were found in TATA box-like
regulatory elements (TA1 to TA4). Mutation of both A1762T and G1764A was found in only one OBIC(SZ6) strain, a lower
frequency than reported in OBIE (44.4%) (21). Synthesis of both pregenomic and 5⬘-terminally extended precore RNA is controlled by the core regulatory elements including ENH2, CURS, and BCP; given the overlapping arrangement of ORFs, every single mutation can thus influence more than one func-tion of the corresponding nucleotide sequence during viral RNA transcription. Mutations in OBI strains in core regula-tory elements that decrease or abolish viral replication, RNA transcription, or production of precore/core protein and se-creted HBe might be responsible for low viral load in OBIB
and OBICstrains.
The mutations of core protein in HBV infection have been reported associated with not only severe liver disease but also low secretion of HBV virions and immune escape epitopes at Th and CTL levels (3, 11, 16). Mutations of OBI core proteins (Fig. 1) showing amino acid substitutions in the CTL epitope clusters 18 to 27, 88 to 96, and 141 to 151 might constitute escape epitopes and contribute to the persistence of HBV infection despite reducing viral fitness (16). The CTL epitope at positions 18 to 27 is restricted by HLA-A2, but the potential impact of substitutions at position 21 and 26 is not known (2). Similarly, CTL epitopes at positions 88 to 96 and 141 to 151 are restricted by HLA-A11 and HLA-Aw68, respectively (9). Sub-stitution L60V (SZ5) was previously described as related to low-level production of HBV virions (16). Mutations of I97(L/T) and P130T were identified in 60% (3/5) and 40% (2/5) OBI strains. I97L was reported responsible for secretion of immature HBV core, but this phenomenon could be offset by P130T mutation (18, 19).
[image:6.585.85.500.69.242.2]The function of X protein could be influenced by the dele-tion, inserdele-tion, and substitutions of core regulatory elements (Table 4) because the X gene overlaps with the enhancer and core promoter regions. Two important domains between aa 52 to 65 and 88 to 154 of HBx have an enhancing effect on HBV transcription and replication (17). At the protein level, substi-tution K130M caused by mutation A1762T in BCP (SZ6) was
FIG. 1. Mutations in core proteins of OBI strains corresponding to immune epitope clusters of CTL, B and Th cells. Epitope clusters were from previous descriptions in HBV infections (3, 16). OBI mutations in the core regions are shown; arrows indicate the epitope clusters. The scale represents the amino acid length of the HBV core protein. The numbers at the ends of the epitopes indicate the start and ending amino acid positions for each epitope.
on May 16, 2020 by guest
http://jcm.asm.org/
observed; other substitutions, V5L, Q87G, and V88(A/N), were involved in 40% (2/5) of OBI strains (SZ19 and SZ6); V88(A/N) and K130M were located in the functional domains of the X protein, but their specific effect on HBV replication is not identified.
In the present study, the diversity of pre-S1 and pre-S2 at the nucleotide and protein levels of OBI genotypes B and C was significantly higher than in control HBsAg⫹ strains (P ⬍
0.0001) (Table 3). However, most mutations appeared to be random substitutions. In contrast, the MHR of OBIB and OBICstrains was essentially wild type, particularly in loops 3 and 4 (Fig. 2D). This pattern does not suggest a major impact of immune selection in genotype B and C OBI strains although only a small number of cases was examined; these findings contrast with findings in European genotype A2 and D but are
rather similar to findings in African genotype A1 or E OBI strains (1, 4).
The relatively high frequency of critical mutations in the precore/core region potentially affecting viral replication and the virtual absence of mutations affecting the pre-S and S proteins suggest that the main mechanism leading to OBI in Shenzhen might be poor viral replication rather than ineffec-tive immune control.
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (973 program numbers 2007CB512901 and 2010CB530204) and Guangdong Research Initiating Funding for talent from overseas.
We thank the staffs of Shenzhen Blood Center and Qingtao Song for their contribution to the study.
We have no conflicts of interest to report.
FIG. 2. Alignments of deduced amino acid sequences of pre-S/S from OBI strains compared to the consensus sequence of a genotype B or C HBsAg⫹ strain. Alignments of pre-S/S were examined for pre-S1(A), pre-S2 (B), S (C), and (D) MHR and “a” region within the S protein. Numbers above the reference sequences indicate epitope clusters (4, 7, 14). Amino acids identified by one-letter code. Dashes indicate identity with the reference consensus sequence for genotype B or C shown on top of each sequence. Amino acid residue K in bold in the reference sequence indicates an HBV serotype-specific amino acid. MHR amino acid mutations in italics may affect serotype or subtype of HBV.
on May 16, 2020 by guest
http://jcm.asm.org/
REFERENCES
1.Allain, J. P., et al.2009. Characterization of occult hepatitis B virus strains in South African blood donors. Hepatology49:1868–1876.
2.Bertoletti, A., et al.1994. Cytotoxic T lymphocyte response to a wild type hepatitis B virus epitope in patients chronically infected by variant viruses carrying substitutions within the epitope. J. Exp. Med.180:933–943. 3.Bozkaya, H., B. Ayola, and A. S. Lok.1996. High rate of mutations in the
hepatitis B core gene during the immune clearance phase of chronic hepatitis B virus infection. Hepatology24:32–37.
4.Candotti, D., et al.2008. Characterization of occult hepatitis B virus from blood donors carrying genotype A2 or genotype D strains. J. Hepatol.49:
537–547.
5.Candotti, D., O. Opare-Sem, H. Rezvan, F. Sarkodie, and J. P. Allain.2006. Molecular and serological characterization of hepatitis B virus in deferred Ghanaian blood donors with and without elevated alanine amino trans-ferase. J. Viral Hepat.13:715–724.
6.Choi, W. C., et al.2009. Window period and occult HBV infection among blood donor population in Hong Kong, p. 114. Abstr. 20th Reg. Cong. Int. Soc. Blood Transfusion, 14 to 18 November 2009, Nagoya, Japan. Interna-tional Society of Blood Transfusion, Amsterdam, Netherlands.
7.Desmond, C. P., A. Bartholomeusz, S. Gaudieri, P. A. Revill, and S. R. Lewin.2008. A systematic review of T-cell epitopes in hepatitis B virus: identification, genotypic variation and relevance to antiviral therapeutics. Antivir. Ther.13:161–175.
8.Guarnieri, M., et al.2006. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J. Virol.
80:587–595.
9.Khakoo, S. I., et al. 2000. Cytotoxic T lymphocyte responses and CTL epitope escape mutation in HBsAg, anti-HBe positive individuals. Gut47:
137–143.
10.Kleinman, S. H., N. Lelie, and M. P. Busch.2009. Infectivity of human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus and risk of transmission by transfusion. Transfusion49:2454–2489.
11.Le Pogam, S., T. T. Yuan, G. K. Sahu, S. Chatterjee, and C. Shih.2000. Low-level secretion of human hepatitis B virus virions caused by two inde-pendent, naturally occurring mutations (P5T and L60V) in the capsid pro-tein. J. Virol.74:9099–9105.
12.Liang, X., et al.2009. Epidemiological serosurvey of hepatitis B in China: declining HBV prevalence due to hepatitis B vaccination. Vaccine27:6550– 6557.
13.Liu, C. J., et al.2009. Hepatitis B virus basal core promoter mutation and DNA load correlate with expression of hepatitis B core antigen in patients with chronic hepatitis B. J. Infect. Dis.199:742–749.
14.Mu, S. C., Y. M. Lin, G. M. Jow, and B. F. Chen.2009. Occult hepatitis B virus infection in hepatitis B vaccinated children in Taiwan. J. Hepatol.
50:264–272.
15.Raimondo, G., et al.2008. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J. Hepatol.49:652–657.
16.Sendi, H., et al.2009. CTL escape mutations of core protein are more frequent in strains of HBeAg negative patients with low levels of HBV DNA. J. Clin. Virol.46:259–264.
17.Tang, H., et al.2005. The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J. Virol.79:5548–5556.
18.Yuan, T. T., and C. Shih.2000. A frequent, naturally occurring mutation (P130T) of human hepatitis B virus core antigen is compensatory for imma-ture secretion phenotype of another frequent variant (I97L). J. Virol.74:
4929–4932.
19.Yuan, T. T., P. C. Tai, and C. Shih.1999. Subtype-independent immature secretion and subtype-dependent replication deficiency of a highly frequent, naturally occurring mutation of human hepatitis B virus core antigen. J. Vi-rol.73:10122–10128.
20.Yuan, Q., et al.2010. Molecular characteristics of occult hepatitis B virus from blood donors in southeast China. J. Clin. Microbiol.48:357–362. 21.Zahn, A., et al.2008. Molecular characterization of occult hepatitis B virus
in genotype E-infected subjects. J. Gen. Virol.89:409–418.