INTRODUCTION
Several types of neurons are generated in the ventral midbrain including DNs, OMNs, RNNs and GABAergic interneurons. They are all associated with essential neuronal functions and are also clinically relevant. DNs are important for physiological functions such as motor control, cognition and reward, and the degeneration of these cells is a hallmark of Parkinson’s disease (Bjorklund and Dunnett, 2007; Haber and Fudge, 1997; Marsden, 1994; Van den Heuvel and Pasterkamp, 2008). A better understanding of DN development in relation to other neuronal subtypes generated in the ventral midbrain could facilitate the directed differentiation of stem cells into DNs for cell replacement therapy for Parkinson’s disease (Arenas, 2010; Hedlund and Perlmann, 2009; Thomas, 2010). However, although several signals and transcriptional regulators that influence the generation of DNs have been identified, it still remains poorly understood how such factors are mechanistically integrated into gene regulatory networks that underlie the precise spatiotemporal specification of cell types in the developing ventral midbrain.
Functionally distinct neuronal subtypes are generated at specific positions and at defined time points during nervous system development. Inductive signals play key roles in neuronal specification by controlling the expression patterns of transcription factors that, in turn, establish distinct neural progenitor domains via cross-repressive transcriptional interactions (Briscoe et al., 2000;
Gunhaga et al., 2003; Muhr et al., 2001; Scholpp et al., 2003). Moreover, these factors specify neuronal subtype identities as progenitors leave the cell cycle and differentiate into post-mitotic neurons (Jessell, 2000). DNs are derived from a pool of neural progenitors located at the ventral midline of the developing midbrain. In the midbrain, signaling by Shh (sonic hedgehog), FGF8 (Fibroblast growth factor 8) and Wnts (Wingless-related MMTV integration site) promote the generation of DNs (Cajanek et al., 2009; Hynes and Rosenthal, 1999; Joksimovic et al., 2009b; McMahon and Bradley, 1990), but these factors also underlie the induction of other ventrally derived neuronal subtypes, such as OMNs, RNNs and GABAergic inhibitory interneurons (Perez-Balaguer et al., 2009; Prakash et al., 2006; Ye et al., 1998). In addition, several transcription factors that influence cell patterning and the differentiation of ventral midbrain neurons have been identified (Andersson et al., 2006b; Ferri et al., 2007; Hasan et al., 2010; Kala et al., 2009; Nakatani et al., 2007; Pattyn et al., 1997; Puelles et al., 2003).
The LIM-homeodomain proteins Lmx1a and Lmx1b are co-expressed in DN progenitors and differentiating DNs and have been shown to mediate key functions in the initial steps of DN fate specification (Andersson et al., 2006b; Smidt et al., 2000). Lmx1a is specifically induced in DN progenitors around embryonic day (E) 9.0 in mice. By contrast, Lmx1b is more broadly expressed in the ventral midbrain at early developmental stages and becomes restricted to DN progenitors only at E10.5 when DNs begin to differentiate (Andersson et al., 2006b). This raises the possibility that Lmx1b is not only important for developing DNs but may also influence specification of other cell types generated at more lateral positions of the ventral midbrain. Moreover, both Lmx1a and Lmx1b are sufficient to induce DNs when ectopically expressed in the midbrain (Andersson et al., 2006b; Lin et al., 2009; Nakatani et al., 2010) or when expressed in differentiating pluripotent stem cells (Andersson et al., 2006b; Friling et al., 2009; Chen et al., 2009). Development 138, 3399-3408 (2011) doi:10.1242/dev.065482
© 2011. Published by The Company of Biologists Ltd
1Karolinska Institutet, Department of Cell and Molecular Biology, von Eulers väg 3, 171 77 Stockholm, Sweden. 2Ludwig Institute for Cancer Research, Nobels väg 3, Karolinska Institutet, 71 77 Stockholm, Sweden. 3Institut de Biologie de l’Ecole Normale Supérieure (IBENS), CNRS UMR8197, INSERM U1024, 75005, Paris, France. 4UMDNJ, Neuroscience and Cell Biology, CABM, 679 Hoes Lane, Piscataway, NJ 08854, USA.
*These authors contributed equally to this work
†Author for correspondence (johan.ericson@ki.se;thomas.perlmann@licr.ki.se)
Accepted 10 June 2011
SUMMARY
The severe disorders associated with a loss or dysfunction of midbrain dopamine neurons (DNs) have intensified research aimed at deciphering developmental programs controlling midbrain development. The homeodomain proteins Lmx1a and Lmx1b are important for the specification of DNs during embryogenesis, but it is unclear to what degree they may mediate redundant or specific functions. Here, we provide evidence showing that DN progenitors in the ventral midbrain can be subdivided into molecularly distinct medial and lateral domains, and these subgroups show different sensitivity to the loss of Lmx1a and Lmx1b. Lmx1a is specifically required for converting non-neuronal floor-plate cells into neuronal DN progenitors, a process that involves the establishment of Notch signaling in ventral midline cells. On the other hand, lateral DN progenitors that do not appear to originate from the floor plate are selectively ablated in Lmx1bmutants. In addition, we also reveal an unanticipated role for Lmx1b in regulating Phox2a expression and the sequential specification of ocular motor neurons (OMNs) and red nucleus neurons (RNNs) from progenitors located lateral to DNs in the midbrain. Our data therefore establish that Lmx1b influences the
differentiation of multiple neuronal subtypes in the ventral midbrain, whereas Lmx1a appears to be exclusively devoted to the differentiation of the DN lineage.
KEY WORDS: Lmx1a/b, Midbrain, Phox2a, Dopamine neuron, Ocular motorneuron, Red nucleus interneuron, Mouse
Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in
ventral midbrain development
Qiaolin Deng1,2, Elisabet Andersson1,*, Eva Hedlund2,*, Zhanna Alekseenko1, Eva Coppola3, Lia Panman2,
James H. Millonig4, Jean-Francois Brunet3, Johan Ericson1,†and Thomas Perlmann1,2,†
D
E
V
E
LO
P
M
E
N
However, the precise roles for Lmx1a and Lmx1b during embryogenesis remain poorly characterized and it is unclear to what degree Lmx1a and Lmx1b mediate redundant or specific activities in midbrain patterning and neuronal fate determination. DN development is affected both in Lmx1b-null mutants and in homozygous dreher(dr/dr) mice carrying an amino acid substitution of a conserved cysteine residue within the first LIM domain of the Lmx1a protein (Ono et al., 2007; Smidt et al., 2000). Thus, these two proteins cannot fully compensate for each other. Importantly, however, although Lmx1a protein expression is abolished in the dorsal neural tube in dr/drembryos, the expression persists in DN progenitors and differentiating DNs within the ventral midbrain and it remains possible that the DN phenotype in dr/drembryos reflects a partial loss of Lmx1a activity (Millonig et al., 2000) (see also below). Additionally, few studies have addressed the requirement for Lmx1b in ventral midbrain development (Smidt et al., 2000) and the precise role for Lmx1b in cell patterning and neuronal fate determination therefore remains unresolved.
To delineate more conclusively the roles of Lmx1a and Lmx1b during ventral midbrain development, we generated two different Lmx1a-null mouse strains where the first two exons were removed, and in one strain replaced with a sequence encoding the enhanced green fluorescence protein (eGFP). Midbrain patterning and specification of DNs were compared in dr/dr, Lmx1a- and Lmx1b-null embryos, and in Lmx1a/Lmx1bcompound mutants. Analysis of these transgenic mouse lines revealed both unique and redundant roles of Lmx1a and Lmx1b in DN generation. Notably, Lmx1a is more important for neurogenesis in the medial DN domain, whereas Lmx1b is more important for the generation of lateral DNs. In addition, our study revealed a broader role for Lmx1b in midbrain development. Phox2a has previously been shown to specify OMNs within the ventral midbrain (Hasan et al., 2010; Nakano et al., 2001; Pattyn et al., 1997). We show here that Lmx1b is required for the expression of Phox2a, and influences the temporal specification of OMNs and RNNs in the ventral midbrain. Thus, Lmx1b and Phox2a are part of a novel gene regulatory network that controls the spatiotemporal generation of DNs, OMNs and RNNs during midbrain development.
MATERIALS AND METHODS Mouse mutants
All experiments on animals have been approved by the Swedish ethical council. The DreherJ(referred to as dr in the text) mouse was obtained from J.H.M.; the Lmx1b-null mouse was obtained from T. Jessell (Columbia University, NY, USA) and M. Mendelsohn (Columbia University, NY, USA); and the Phox2a-lacZmouse has been described by Jacob et al. (Jacob et al., 2000). All lines were maintained on a C57BL/6J background. Genotyping was carried out as previously described (Chen et al., 1998; Millonig et al., 2000). The Lmx1aeGFP/+mouse was generated by
the Institut Clinique de la Souris, France. The cloning strategy is shown in Fig. S1A in the supplementary material. In short, the genomic 3⬘and 5⬘
Lmx1a arms were isolated using PCR, subcloned and verified by sequencing. The 3 kb 5⬘arm was cloned into a lox-neo-lox (lNeol) vector using FseI/PacI. The enhanced green fluorescent protein (eGFP) and the 4.5 kb 3⬘arm were added using PacI/SgrAI and SbfI/SfiI, respectively. Homologous recombination was confirmed using Southern blot hybridization with 5⬘and 3⬘external probes. Exon 1, intron 1, exon 2 and part of intron 2 were removed. The Lmx1a conditional knockout mouse (Lmx1acko/+) was generated in our laboratory. The cloning strategy is shown
in Fig. S1B in the supplementary material. In short, the genomic 3⬘and 5⬘
Lmx1a arms were amplified by PCR from RPCI-21 PAC library and verified by sequencing. Single loxP sites was inserted upstream of exon 1 together with an extra BglII site used for screening. The lNeol cassette was inserted 288 nucleotides downstream of exon 2, in intron 2, with an extra
ApaLI site for screening. The initial screen was performed by long-range PCR (internal neo primer and 3⬘external primer) and the final validation was carried out using Southern blot hybridization with a 5⬘and 3⬘external probe (see Fig. S1Ba,b in the supplementary material). Chimeric mice were generated by KCTT, Karolinska Institutet, Sweden. Germline transmission was verified by Southern blot and PCR. This mouse line was maintained as a homozygous line (Lmx1acko/cko) and showed a behavior that was indistinguishable from wild-type mice. To delete exon 1, intron 1, exon 2 and part of intron 2 of the Lmx1agene, the Lmx1acko/ckomouse line was crossed with a CMV-Cre mouse line and the resulting mouse line was termed Lmx1a+/–. This line was maintained as heterozygote and
subsequently intercrossed to generate Lmx1a-null mice (Lmx1a–/–).
Immunohistochemistry and in situ hybridization
The following antibodies were used: mouse Foxa2, Lim1/2, Nkx2.2, Shh and Nkx6.1 (Developmental Studies Hybridoma Bank); mouse TH and Brn3a (Chemicon); mouse Ngn2 (a gift from D. Anderson, California Institute of Technology, CA, USA); and mouse Gata3 (Santa Cruz); rabbit GFP (Molecular Probes), rabbit Lmx1a (a gift from M. German, UCSF, CA, USA), rabbit Nurr1 (Santa Cruz), rabbit TH (PelFreeze), rabbit Phox2a (prepared by J.-F.B.) and rabbit Corin (a gift from M. Parmar, Lund University, Lund, Sweden); guinea pig Lmx1a, Lmx1b and Pitx3 (prepared by J.E.), guinea pig Isl1/2 (T. Jessell) and guinea pig Phox2b (prepared by J.E.); goat -galactosidase (Biogenesis) and goat GFP (Abcam). In situ hybridization on sections was carried out essentially as previously described (Briscoe et al., 2000) using probes for Wnt1(Puelles et al., 2003), Dll1
(Lindsell et al., 1996), Ngn2(Fode et al., 1998), Hes5(Akazawa et al., 1992),
Tcf12(Riken clone AK078415), Drd2(Geneservice I.M.A.G.E ID 13489) and Sim1(Pierani et al., 1999).
Quantification of DNs in Lmx1a and Lmx1b mutant mice
For evaluation of the importance of Lmx1a and Lmx1b for DN generation in the ventral midbrain, DN markers were counted at E11.5 (Nurr1), E13.5 (Nurr1 and Pitx3) and P0 (TH) in wild-type, dr/dr, Lmx1a-null, Lmx1b -null and/or Lmx1a/bcompound mutant mice. At E11.5 and E13.5, all Nurr1+and/or Pitx3+neurons in the ventral midbrain were counted in four
consecutive sections/embryo (four embryos per genotype). At P0, TH+
neurons in the midbrain were counted unilaterally with a subdivision into two regions, 0-250 M and 250-500 M from the midline. One out of five sections (12 m sections) were counted across four sections/animal. Four wild-type and one heterozygous (included in the wild-type group) and five
Lmx1aeGFP/eGFPanimals were counted. All counts were performed in a double-blind fashion.
DNA constructs and in ovo electroporation
cDNAs encoding mouse Lmx1a (Riken; AK044944), Lmx1b (Q. Ma) and
Lmx1adr (mimicking dr mutation with C82Y) in RCASBP were electroporated (Briscoe et al., 2000) at 1.5 g/l into the midbrain of HH stage 10-12 chick embryos. Chick embryos were analyzed 84 hours after electroporation.
cDNAs encoding mouse Lmx1a in CAGG:iresGFP was electroporated at 0.8 g/l into the midbrain of HH stage 10-12 chick embryos. Chick embryos were analyzed 48 hours after electroporation.
Optical density measurements
Optical density of Lmx1b and GFP was measured in DNs that were transfected with CAGG-Lmx1a:iresGFP construct using ImageJ (http://imagej.nih.gov/ij/). Confocal images used for the optical density measurements were carefully captured at a setting where all cells displayed signal intensity below the saturation level. A few extreme high level of GFP+cells were excluded from the measurements. The optical density
values were normalized by the medium optical density for each embryos (n7) and the relationship depicted in a scatter plot.
Statistical analysis
P0 experimental data was analyzed using Student’s t-test and ANOVA. InStat3 software (GraphPad software) was used for the statistical analyses. Experimental data from other different embryonic stages was analyzed
using mean±s.e.m. values.
D
E
V
E
LO
P
M
E
N
RESULTS
Generation of Lmx1a-null mice
To determine whether the reduction of DNs observed in dr/drmice reflects a partial or complete loss of Lmx1a activity (Ono et al., 2007), we generated two new Lmx1a mutant mouse strains (Lmx1aeGFP and Lmx1acko) (see Fig. S1 in the supplementary material). In Lmx1aeGFP/+heterozygous mice, exons 1 and 2 in one Lmx1a gene copy were replaced with a sequence encoding eGFP (see Fig. S1A in the supplementary material). In heterozygous Lmx1aeGFP/+ embryos, expression of the eGFP reporter recapitulated the expression profile of Lmx1a in the ventral midbrain, as well as in other regions in which Lmx1ais expressed during embryogenesis (Fig. 1A; data not shown). In heterozygous Lmx1acko/+mice, LoxP sites were introduced in one Lmx1agene copy allowing conditional deletion of exons 1 and 2 (see Fig. S1B in the supplementary material). Lmx1a-null mutant embryos (termed Lmx1a–/–) were generated by using these mice in crosses with ‘deleter’ mice expressing the Cre-recombinase under the
CMV promoter (Su et al., 2002). Importantly, in contrast to wild-type and dr/drmice, no Lmx1a protein was detected in the ventral midbrain of either Lmx1aeGFP/eGFPor Lmx1a–/–embryos at E11.5 (Fig. 1B). Thus, Lmx1aeGFP/eGFP and Lmx1a–/– mice represent Lmx1a-null mutants.
We next examined the generation of DNs in the ventral midbrain at E11.5 and E13.5 in the different Lmx1a mutant backgrounds. In both Lmx1aeGFP/eGFPand Lmx1a–/–embryos, the number of DNs was significantly reduced, as determined by analyzing the co-expression of Nurr1 and Pitx3 at E13.5 (Fig. 1C). Quantification of Nurr1 and Pitx3 expressing cells showed that the partial loss of DNs was similar in the different mutant backgrounds (~60% at E11.5 and ~30-40% at E13.5; Fig. 1D). Thus, these data reveal that Lmx1a is not absolutely required for the specification of DNs and also suggest that the drmutation results in a complete loss of Lmx1a function. This conclusion is supported by experiments showing that the ability of Lmx1a to induce ectopic DNs in the chick midbrain was completely abolished after introducing the dr mutation in the Lmx1a expression construct (Fig. 1E; data not shown).
Non-redundant functions of Lmx1a and Lmx1b in DN fate specification
Lmx1a and Lmx1b are co-expressed in DN progenitors and differentiating DNs in wild-type embryos at E11.5 (Fig. 2A), although Lmx1b is more broadly expressed in the ventral midbrain at early developmental stages (Andersson et al., 2006b) (see below). Thus, Lmx1b may functionally compensate for the loss of Lmx1a. The DN progenitor domain in Lmx1aeGFP/eGFPembryos was similar in size to that of wild-type embryos, as determined by Lmx1b and Foxa2 expression at E11.5 (Fig. 2A,B,D,E). Despite the normal size of the DN progenitor domain in Lmx1aeGFP/eGFP embryos, generation of post-mitotic DNs was limited to lateral positions and essentially no Nurr1+/TH+cells could be detected at the ventral midline of the midbrain at E11.5 (Fig. 2E,H) (Ono et al., 2007). Moreover, the expression of Lmx1b was significantly upregulated in the DN progenitor domain of Lmx1aeGFP/eGFP embryos (Fig. 2B). The upregulated expression was seen in the entire Lmx1a-postive progenitor domain as determined by double staining for both Lmx1b and Corin (see Fig. S2 in the supplementary material). Thus, these results identify a unique requirement for Lmx1a in the control of DN neurogenesis in the medial part of the progenitor domain. In addition, these results show that the elevated expression of Lmx1b cannot compensate for the loss of Lmx1a function in the medial DN progenitor domain.
[image:3.612.52.296.277.579.2]In parallel experiments, we analyzed the ventral midbrain in Lmx1bmutant (Lmx1b–/–) embryos (Chen et al., 1998). Lmx1b is known to be sufficient to induce Lmx1aexpression (Chizhikov and Millen, 2004; Nakatani et al., 2010), a result that we confirmed by introducing an Lmx1bexpression construct into the chick neural tube (see Fig. S3 in the supplementary material). However, DN progenitors expressing Lmx1a and Foxa2 could be detected at the ventral midline of Lmx1b–/–embryos showing that Lmx1bis not absolutely required for the induction of Lmx1aexpression in this region (Fig. 2C,F). Moreover, although the size of the DN progenitor domain in Lmx1b–/–was substantially reduced compared with wild-type or Lmx1aeGFP/eGFPembryos, a significant number of medially located neurons expressed Nurr1 and TH in Lmx1b–/– embryos at E11.5 (Fig. 2F,I). These data raised the possibility that loss of Lmx1b primarily affects the establishment of the lateral part of the DN progenitor domain. We therefore analyzed additional markers that could distinguish between the medial and lateral DN Fig. 1.dr/dr, Lmx1aeGFP/eGFPand Lmx1a–/–mice show a similar DN
phenotype in the ventral midbrain.(A)In E12.5 Lmx1aeGFP/+ embryos, eGFP is expressed in discrete locations, including the ventral midbrain (Vm), roof plate and cortical hemisphere (BF, bright field). Sections of ventral midbrain (bottom) show that eGFP is co-expressed with Lmx1a. (B)Transverse sections of ventral midbrain at E11.5 show that Lmx1a protein is undetectable in Lmx1aeGFP/eGFPand Lmx1a–/–, but is present in dr/drand wild type. (C)At E13.5, Nurr1+/Pitx3+DNs were reduced in dr/dr, Lmx1aeGFP/eGFPand Lmx1a–/–compared with wild type. (D)Analysis of Nurr1+DNs at E11.5 and E13.5, and Pitx3+DNs at E13.5. Values are presented as mean±s.d., n4. (E)In chick electroporation, Lmx1awtand Lmx1bwtinduce Nurr1+cells 84 hours post-transfection, but not the eGFP control or a construct carrying the
drmutation of Lmx1a(Lmx1adr). Endogenous post-mitotic DNs are
indicated by the yellow dotted line.
D
E
V
E
LO
P
M
E
N
progenitor domains. Wnt1 was selectively expressed in lateral DN progenitors at E11.5 (Fig. 2M) (McMahon and Bradley, 1990; Prakash et al., 2006). In addition, the dopamine receptor D2 (Drd2 or D2R) was restricted to lateral DNs at E13.5 (Fig. 2S) (Kim et al., 2006). Interestingly, both of these lateral markers were almost completely lost in Lmx1b–/– embryos while remaining largely unaffected in Lmx1aeGFP/eGFPembryos (Fig. 2N,O,T,U). Moreover, the expression of Corin, which selectively defines ventral midline cells, was retained in both Lmx1aeGFP/eGFPand Lmx1b–/–embryos (Fig. 2J-L). However, lateral DN progenitors residing in between the expression domains of Corin and Nkx6.1 were abolished in Lmx1bbut not Lmx1amutants at E11.5 (Fig. 2J-L). Thus, these data show that the DN progenitor domain can be subdivided into at least two distinct (medial and lateral) zones, and suggest that Lmx1a is selectively required for the specification of DNs in the medial DN progenitor domain, whereas Lmx1b is necessary for the establishment of the lateral DN progenitor domain (Fig. 2P-R). Nevertheless, it is noteworthy that differentiation of medially derived DNs is also affected by the loss of Lmx1b, as the majority of Nurr1+DNs detected in Lmx1bmutants fail to initiate expression of late DN markers including TH and Pitx3 (Fig. 2X; data not shown) (Smidt et al., 2000).
Our comparison of Lmx1aand Lmx1bsingle mutants revealed unique roles for these two proteins in DN progenitors. To explore whether Lmx1a and Lmx1b also mediate redundant activities, we analyzed the generation of DNs in different Lmx1a/Lmx1b compound mutant backgrounds at E13.5. Embryos heterozygous both for Lmx1aand Lmx1bexhibit a ~20-25% reduction in the number of Nurr1- and Pitx3-expressing cells compared with wild-type and single Lmx1aor Lmx1bheterozygous embryos (Fig. 2Y). In addition, a dramatic reduction in the number of DNs was evident when Lmx1a-null mutant embryos carried one mutant Lmx1ballele (~70-75% loss of Nurr1- and Pitx3-expressing cells compared with a ~30-40% reduction in Lmx1ahomozygous embryos) (Fig. 2Y). Owing to the early lethality, presumably reflecting early developmental functions of Lmx1a and Lmx1b, we were unable to obtain E13.5 embryos that were null for both genes. However, the Lmx1agene dose-dependent reduction of DNs strongly indicates that the combined and partly redundant activities of Lmx1a and Lmx1b are essential for specification of all DNs in the ventral midbrain.
[image:4.612.52.296.56.526.2]Lmx1a ablation delays induction of the Notch pathway within the medial ventral midbrain Midbrain DNs are unique in that they are derived from floor-plate cells. By contrast, floor-plate cells located at more caudal axial levels do not generate any neurons, indicating that ventral midbrain floor-plate cells must acquire neurogenic properties during their development (Joksimovic et al., 2009b; Ono et al., 2007). Lmx1a is important for the induction of Ngn2 (Ono et al., 2007), a proneural bHLH protein that promotes the generation of post-mitotic DNs (Andersson et al., 2006a; Kele et al., 2006), and may thus regulate the switch from non-neuronal floor plate cells to neuronal DN progenitors. Additional genes implicated in the regulation of neurogenesis were analyzed in Lmx1aeGFP/eGFP embryos, including components of the Notch signaling pathway. Interestingly, along with Ngn2, expression of the Notch ligand Dll1 as well as of the bHLH proteins Hes5 and Tcf12 was selectively lost in the medial part of the DN progenitor domain in these embryos at E11.5 (Fig. 3A-H). Hes5 operates downstream of Dll1 in the Notch pathway and functions to suppress the expression of proneural bHLH genes (Ohtsuka et al., 1999). By contrast, Tcf12 Fig. 2. Characterization of the DN domain in Lmx1aeGFP/eGFPand
Lmx1b–/–mice.Transverse sections of the ventral midbrain at E11.5 and
E13.5, comparing expression patterns in wild type, Lmx1aeGFP/eGFPand
Lmx1b–/–. (A,B)Lmx1bexpression remained unchanged, even enhanced, in DN progenitors in Lmx1aeGFP/eGFP(B) compared with wild type (A). (C)In
Lmx1b–/–, the Lmx1adomain became smaller. (D-I)There was a specific medial loss of DNs in Lmx1aeGFP/eGFPshown by the lack of Nurr1 (E,H) and TH (H) which is not seen in wild type (D,G) and Lmx1b–/–(F,I). The medial region is marked by white bracket. (J-L)Corin expression showed that the medial domain was maintained in both mutants (K,L). By contrast, the lateral domain was lost in Lmx1b–/–(L) but not in Lmx1aGFP/GFP(K), as indicated by the loss of the gap between Corin and Nkx6.1 seen in the wild type (J). (M-O,S-U) In situ hybridization of Wnt1and Drd2in wild type (M,S), Lmx1aeGFP/eGFP(N,T) and Lmx1b–/–(O,U) showed that Wnt1 andDrd2expression were lost in Lmx1b–/–, but remained unchanged in
Lmx1aeGFP/eGFP. (P-R)Schematic model of the medial (DNM) and lateral (DNL) DN domains at E11.5. (V-X)At E13.5, Nurr1+DNs in Lmx1b–/–failed to initiate mature DN markers (X) compared with wild type (V) and
Lmx1aeGFP/eGFP(W). (Y)The percentage of Nurr1+or Pitx3+DNs of wild type in different genotypes of Lmx1a/Lmx1bcompound mutant mice.
Values are presented as mean±s.d., n4.
D
E
V
E
LO
P
M
E
N
functions to promote transcription of neuronal genes by heterodimerizing with proneural bHLH proteins (Neuman et al., 1993). Thus, Lmx1a appears to promote the expression of several genes implicated in the Notch signaling pathway, presumably facilitating Notch signaling and establishing neuronal potential in floor plate cells at the ventral midline.
The induction of Ngn2 and concomitant loss of Shh expression that occurs in midline cells at around E10.5-E11.5 are indicative of a conversion of floor-plate cells into DN progenitors (Fig. 3I,L) (Andersson et al., 2006b; Joksimovic et al., 2009b). These regulatory events occurred normally in Lmx1b–/–mice supporting that loss of Lmx1b primarily affects lateral DN progenitors, which are not derived from floor-plate cells (Fig. 3K,N). By contrast, loss of Ngn2 expression accompanied by maintained Shh expression in the midline was detected in Lmx1aeGFP/eGFPembryos at E11.5 (Fig. 3J,M). However, from E12.5 and onwards, Shh became suppressed and Ngn2, as well as Notch signaling-related genes, became activated in these embryos, which is consistent with the previously observed partial restoration of DNs in dr/drmice over time (Fig. 3O-T) (Ono et al., 2007). Thus, the initial loss of DNs in Lmx1aeGFP/eGFP embryos most probably reflects a prolonged maintenance of floor-plate properties and delayed neurogenesis at the ventral midline. Interestingly, however, a modest but significant loss of DNs persists in early postnatal Lmx1aeGFP/eGFPmice within the ventral tegmental area (VTA) (~20% reduction within the specified region) demonstrating that the medial delay in neurogenesis cannot be fully compensated for at later developmental stages (Fig. 4A,B).
Lmx1b controls the generation of OMNs and RNNs located lateral to midbrain DNs
Studies of Lmx1a and Lmx1b in midbrain development have primarily focused on the generation of DNs. However, in contrast to Lmx1a, Lmx1b is broadly expressed in the ventral midbrain at early developmental stages (Andersson et al., 2006b). We, therefore, examined the expression of Lmx1b relative to other cell fate-determining transcription factors expressed in midbrain
[image:5.612.51.356.59.311.2]progenitors and examined how the generation of additional neuronal populations was affected in Lmx1aand Lmx1bmutants. Based on the expression of transcription factors, at least three molecularly distinct progenitor domains can be defined in the ventral-most midbrain at E10.5. First, Lmx1a+cells define DN progenitors at the ventral midline. Second, OMNs are generated immediately lateral to the Lmx1a+/Lmx1b+DN progenitor domain from cells that express Phox2a, Sim1 and Nkx6.1 (Fig. 5C,D; see also Fig. 6A,D,G,J). RNNs are also generated from this region, although the precise spatiotemporal origin of these neurons remains
Fig. 3. Lmx1aeGFP/eGFPmice showed defects in
neurogenesis in the ventral medial region. (A-H)Transverse sections of wild-type, Lmx1aeGFP/eGFP and Lmx1b–/–embryos at E11.5 and E12.5. In situ hybridization of Ngn2, Tcf12, Dll1and Hes5RNAs at E11.5 showed a specific medial loss in Lmx1aeGFP/eGFP (B,D,F,H) compared with wild type (A,C,E,G). (I-N)At E11.5, in Lmx1b–/–, the expression of Ngn2 and Shh (K,N) was similar to the wild type (I,L), in contrast to
Lmx1aeGFP/eGFP(J,M). (O-T)At E12.5, the expression pattern of Hes5, Ngn2and Shh was restored in the
Lmx1aeGFP/eGFP. Red brackets indicate the ventral medial region; broken red lines indicate the DN domain.
Fig. 4. Postnatal Lmx1aeGFP/eGFPmice show a significant loss of
VTA DNs.DNs in the VTA of wild-type (n5) and Lmx1aeGFP/eGFP(n5) mice were quantified at P0. (A)The VTA of 500m is shown in the picture and divided into regions of 0-250m and 250-500m from the ventral midline (broken red line indicating 250m) and all TH neurons within these subregions were subsequently counted unilaterally. (B)A ~20% reduction in the number of TH+neurons within the 250-500m region of the VTA was detected (0-250m: wild type, 124.3±12.7;
Lmx1aeGFP/eGFP, 111.6±12.2; 250-500m: wild type, 184.8±9.3;
Lmx1aeGFP/eGFP, 147.9±9.2) (P0.01 at 250-500m, analyzed by
Student’s t-test).
D
E
V
E
LO
P
M
E
N
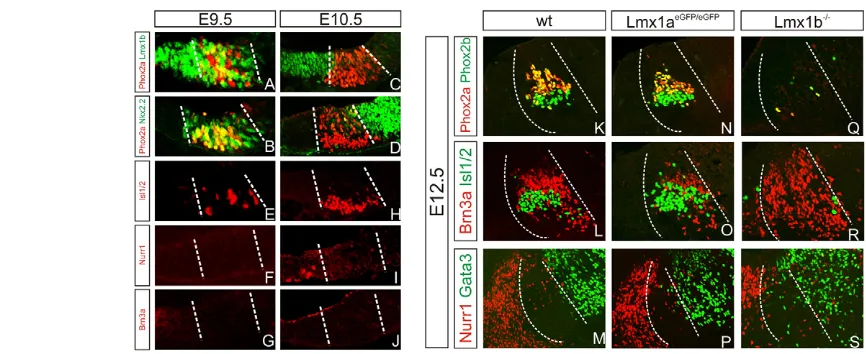
[image:5.612.331.547.471.618.2]uncertain (Prakash et al., 2009). Third, progenitors located lateral to the Phox2a+OMN progenitors express Nkx2.2 and differentiate into Gata2/3+GABAergic neurons (Fig. 5D,M) (Kala et al., 2009; Nakatani et al., 2007). At E9.5, many lateral Lmx1b+cells co-expressed Phox2a and Nkx2.2, and the expression of these genes did not segregate into distinct expression domains until E10.5 (Fig. 5A-D). As Isl1/2+OMNs, but not DNs and RNNs, have been generated already by E9.5, these data strongly suggest that at least early born OMNs originate from progenitors that express Phox2a, Lmx1b and Nkx2.2 (Fig. 5E-J). Analysis of neuronal subtype-specific markers at E12.5 indicated that the generation of OMNs, RNNs and GABAergic neurons was unaffected by the loss of Lmx1a(Fig. 5K-P). By contrast, OMNs were almost completely lost in Lmx1b mutants at E12.5 and this phenotype was accompanied by a significant increase in the number of Brn3a+ RNNs (Fig. 5Q,R). As in controls, these cells were located in between Nurr1+DNs and Gata3+ GABAergic neurons (Fig. 5M,S).
Additional markers were analyzed at E10.5-E11.5 (Fig. 6). This analysis showed that the patterned expression of Lmx1a, Nkx6.1 and Nkx2.2 remained largely unaffected in Lmx1bmutants at E10.5 (Fig. 6B,E,H). However, Phox2a expression was almost completely lost in Lmx1bmutants at E10.5, whereas the expression of Sim1 was upregulated and had further extended ectopically into the Lmx1a+progenitor domain at the ventral midline (Fig. 6E,K). Thus, these data strongly suggest that OMNs and RNNs are derived from the same progenitor domain but at different time points, and that Lmx1b is necessary for the generation of OMNs and for the suppression of RNNs at early developmental stages. Notably, and in support of this idea, an extensive overproduction of Brn3a+and Lim1/2+RNNs was observed in Lmx1bmutants already by E11.25, a stage when only few RNNs could be detected in wild-type controls (Fig. 6M,N,P,Q). Scattered Lim1/2+ and Brn3a+ cells were also found intermingled with Lmx1a+ and Nurr1+ cells in Lmx1b mutants, indicating that Lmx1b also functions to suppress the generation of RNNs within the DN progenitor domain (Fig. 6N,Q).
Because the analysis showed that Lmx1b operates upstream of Phox2a in the specification of OMN fate, we speculated that the temporal mis-specification of RNNs in Lmx1bmutants reflected the downregulation of Phox2a. To directly test this idea, Phox2amutant embryos (Jacob et al., 2000) were analyzed. Interestingly, although spatial patterning appeared normal (Fig. 6C,F,I,L), loss of Phox2a resulted in a similar phenotype to that observed in Lmx1bmutants: the loss of OMNs and a striking overproduction of Lim1/2+and Brn3a+RNNs at E11.25 (Fig. 6O,R). The coding exons are replaced by the lacZreporter gene in this Phox2amutant allele, which allows short-term tracing of Phox2a-expressing cells. Many Brn3a+and Lim1/2+cells co-expressed -gal in Phox2alacZ/lacZembryos at E11.5 while lacZexpression appeared exclusively confined to the Isl1/2+ OMN lineage in Phox2alacZ/+control embryos (Fig. 6S-X). Thus, cells fated to generate OMNs differentiate into RNNs in the absence of Phox2a function. Taken together, these data establish novel roles for Lmx1b and Phox2a in controlling the sequential generation of OMNs and RNNs from a common progenitor pool in the ventral midbrain. However, it is notable that Lmx1b may also function partly independently of Phox2a in this process as the loss of Lmx1b, but not of Phox2a, increased Sim1 expression and induced ectopic production of RNNs also in the DN progenitor domain at the ventral midline (Fig. 6K,L,N,O,Q,R).
DISCUSSION
[image:6.612.57.490.58.235.2]As Lmx1aand Lmx1bcan activate one another when their expression is forced in progenitors or differentiating DNs, it has remained unclear to what extent Lmx1a and Lmx1b mediate redundant and/or unique activities during ventral midbrain development (Andersson et al., 2006b; Chung et al., 2009; Nakatani et al., 2010). In addition, the role of Lmx1b in ventral midbrain patterning has remained poorly understood (Guo et al., 2007; Smidt et al., 2000). Thus, our current analysis of Lmx1a and Lmx1b mutant mouse embryos provide important new insight into unique and redundant activities for Lmx1a and Lmx1b in DN fate specification, and have revealed entirely unexpected roles of Lmx1b and Phox2a in the temporal regulation of OMN and RNN generation.
Fig. 5. Characterization of lateral domains in the ventral midbrain.(A)Transverse sections of ventral midbrain at E9.5, E10.5 and E12.5. At E9.5, Lmx1b is co-expressed with Phox2a+progenitors lateral of the DN progenitor domain. (B)Nkx2.2 is co-expressed with Phox2a. (C,D)At E10.5, Lmx1b (C), Phox2a (C,D) and Nkx2.2 (D) expression are segregated into three defined domains. (E-J)Isl1/2+OMNs are detected from E9.5 (E,H); Nurr1+DNs are detected from E10.5 (F,I); Brn3a+RNNs are not detected at these time points (G,J). (K-M,Q-S) Lmx1b–/–mutants showed a loss of OMNs (Q,R), an increase of RNNs (R) and largely unaffected GABAergic interneurons (S) when compared with wild type (K-M). (N-P)Lmx1aeGFP/eGFP mutants did not show any effects on the lateral populations.
D
E
V
E
LO
P
M
E
N
Our analysis of Lmx1a- and Lmx1b-null mutant mice confirms that Lmx1a is not absolutely required for the specification of DNs. However, ablation of Lmx1aand Lmx1balleles within the same embryos strongly indicated that the combined Lmx1a/b activities is essential for generation of all DNs. Moreover, this study also
revealed specific requirements for Lmx1a and Lmx1b in medial and lateral DN progenitors, respectively. Importantly, these data establish that the DN progenitor domain is not a homogenous population of cells, but can be subdivided into distinct medial and lateral subpopulations that can be distinguished based on expression of subtype-specific markers and on their differential dependency on Lmx1a and Lmx1b for their early development (Fig. 7). It is notable that Lmx1a previously have been implicated as a key regulator of Wnt1 expression in the midbrain (Chung et al., 2009). However, we found that the expression of Wnt1 was largely unaffected in the midbrain of Lmx1a mutants, but was almost completely abolished in Lmx1bmutants. Consistent with the early role for Lmx1b in establishing the isthmus at the midbrain/hindbrain boundary (Guo et al., 2007), our loss-of-function analysis also indicates that Lmx1b, in contrast to Lmx1a, is a crucial regulator of Wnt1 expression in midbrain DN progenitors at later developmental stages.
[image:7.612.54.295.59.465.2]The floor plate is restricted to the extreme ventral midline at most axial levels of the neural tube, but has been proposed to be broader and encompass the entire DN progenitor domain in the midbrain (Andersson et al., 2006b; Ono et al., 2007; Placzek and Briscoe, 2005). This conclusion is primarily based on studies showing that floor-plate cells and DN progenitors express several common genes, including several of those identified as floor-plate markers in the spinal cord (Ericson et al., 1995; Sasaki and Hogan, 1994). However, the molecular differences between medial and lateral DN progenitors identified here, and the finding that Wnt1+ cells are largely unaffected in Lmx1amutants, suggest that only medially derived DNs originate from bona fide floor plate cells. Like most other progenitors in the neural tube, laterally derived DNs are likely to be derived from progenitors that have already acquired their neurogenic potential at early developmental stages (Lek et al., 2010; Placzek and Briscoe, 2005). In contrast to Lmx1b, Lmx1a has a unique role in mediating the conversion of floor-plate cells into neurogenic DN progenitors, and consistent with a previous analysis of dr/drmice, Ngn2expression is initially diminished in the medial DN progenitor domain in Lmx1aeGFP/eGFP mice (Ono et al., 2007). However, in addition to Ngn2, several additional genes implicated in the Notch signaling pathway are initially lost at the midline of Lmx1aeGFP/eGFP and the downregulation of Shh expression was delayed. It is therefore conceivable that Lmx1a not only triggers cell cycle exit and neuronal differentiation simply by activating Ngn2, but also facilitates the establishment of functional Notch signaling and lateral inhibition at the ventral midline, which thereby provides neuronal potential to floor-plate cells (Fig. 7). However, additional activities, including Wnt signaling (Prakash et al., 2006), are also likely to contribute to this process as the requirement for Lmx1a in midline cells is limited to early developmental stages and neurogenesis recovers over time in the midbrain of Lmx1amutants. Functionally distinct DNs are topologically organized in defined areas of the midbrain. DNs of the VTA, which are involved in motivational and cognitive functions, occupy the ventral midline, whereas cells of the substantia nigra pars compacta (SNpc), which are implicated in motor control, are located in more lateral positions (Bjorklund and Dunnett, 2007; Haber and Fudge, 1997; Van den Heuvel and Pasterkamp, 2008). It remains unclear how distinct DN subtypes are established during development, but it is conceivable that the molecularly distinct medial and lateral DN progenitors contribute to discrete DN subtype populations. We found that the Drd2 is a marker of lateral DNs at E13.5 and that these cells are missing in Lmx1b-null, but spared in Lmx1a-null, Fig. 6. Lmx1b is involved in the patterning and generation of
lateral neuronal subtypes in the ventral midbrain. (A-L)Transverse sections of ventral midbrain at E10.5, E11.25 and E11.5. At E10.5, three progenitor domains could be identified in both Lmx1b–/–and
Phox2alacZ/lacZ, as defined by combined expression of Lmx1a, Lmx1b, Phox2a, Nkx6.1, Nkx2.2 and Sim1 (broken lines). In Lmx1b–/–, Phox2a+ progenitors were lost (E) and the Sim1+domain was expanded into the DN domain (K). By contrast, in Phox2alacZ/lacZ, the Sim1+domain remained unchanged (L). (M,P)At E11.25, Isl1/2+OMNs (broken circle), Brn3a+RNNs and Lim1/2+cells are expressed dorsal to the DN domain (broken line) in wild type. (N,Q)In Lmx1b–/–, the majority of Isl1/2+ OMNs was lost, whereas the number of Brn3a+RNNs and Lim1/2+cells was increased and encroached into the DN domain. (O,R)Isl1/2+OMNs were lost in Phox2alacZ/lacZaccompanied by an increase of Brn3a+RNNs and Lim1+cells within their normal domain. (S-U)-Gal staining of E11.5 Phox2alacZ/+showed that the Isl1/2+OMNs (S), but not Brn3a+ RNNs (T) and Lim1/2+cells (U), are derived from Phox2a+progenitors. (V-X)In Phox2alacZ/lacZ, -gal+/Brn3a+(W) and -gal+/Lim1/2+(X) cells could be detected, but not Isl1/2+cells (V).
D
E
V
E
LO
P
M
E
N
mutants. These findings indicate that lateral DNs detected at this stage are generated from lateral DN progenitors. Thus, although additional lineage-tracing experiments are necessary, the distinct medial and lateral progenitor domains defined in this study, together with previous cell-tracing experiments showing that ‘young’ floor-plate cells primarily generate VTA DNs, begin to suggest a lineage relationship between DN progenitor populations with defined molecular properties (Joksimovic et al., 2009a). In this context, it is interesting that we observed a small but significant decrease in the number of VTA neurons that persists in newborn Lmx1a-null mice.
Importantly, our analysis shows that Lmx1b is not only devoted to DN specification in the midbrain as the loss of Lmx1b also affects the generation of OMNs and RNNs located lateral to DNs. There is a dramatic reduction of Phox2a expression and a concomitant loss of OMNs in the ventral midbrain of Lmx1b-null mutants, suggesting that Lmx1b operates upstream of Phox2a in OMN fate specification (Fig. 7). The loss of OMNs does not simply reflect a failure to establish defined ventral progenitor domains, as determined by the expression pattern of Lmx1a, Sim1 and Nkx2.2 in Lmx1b-null mutants. Instead, OMNs and RNNs originate from the same ventral progenitor pool, but at sequential developmental stages, and Lmx1b controls this process by promoting the generation of OMNs and suppressing RNNs at early time points. Moreover, lacZ-lineage analysis in Phox2amutant
cells provided direct evidence that these neuronal subtypes have a common origin. The premature induction and extensive overproduction of RNNs observed in Phox2amutants also indicate that a primary role of Lmx1b in this process is to trigger Phox2a expression, which subsequently specifies OMNs while suppressing RNN fate within the OMN domain (Fig. 7) (Hasan et al., 2010).
It remains unclear how Lmx1b mediates this activity, but the broad expression pattern of Lmx1b at an early developmental stage raises the possibility that Lmx1b directly induces Phox2a transcription in OMN progenitors at early stages of midbrain development. However, as Lmx1b is also required for Wnt1 expression in DN progenitors, we cannot exclude that a non-cell autonomous role for Lmx1b contributes to the induction of Phox2a. It is notable that Phox2b, which is highly related to Phox2a, can act as a temporal cell fate determinant by promoting the generation of visceral motoneurons and suppressing serotonergic (5HT) neurons in the ventral hindbrain (Pattyn et al., 1997; Pattyn et al., 2003). A unifying role of these two factors may thus be to promote early born and suppress late-born neurons in the ventral neural tube, an idea that raises the interesting possibility that Phox2a and Phox2b also control temporal cell fate decisions at other sites in the developing CNS and/or peripheral nervous system where these transcription factors are known to be expressed (Pattyn et al., 2000; Pattyn et al., 2003).
Furthermore, our data indicate that Lmx1b can suppress RNN fate in a Phox2a-independent manner as ectopic RNNs are also generated within the DN progenitor domain in Lmx1b-mutant embryos. In normal development, Sim1 and Phox2a are co-expressed in OMN progenitors; however, Phox2a expression is almost abolished in Lmx1bmutants, whereas Sim1 expression is ectopically upregulated in presumptive DN progenitors at the ventral midline. As the expression of Sim1 is unaffected in Phox2amutants, and the premature generation of RNNs in these embryos is limited to the OMN progenitor domain, it seems plausible that Sim1 functions as a determinant of RNNs and that Lmx1b maintains the integrity of DN progenitors by suppressing Sim1 in ventral midline progenitors (Fig. 7). Lmx1b would thus promote DN fate and concomitantly limit the probability that DN progenitors adopt an aberrant RNN differentiation program. However, in more lateral OMN progenitors where Sim1 is normally expressed, it can be predicted that the cell fate-determining activity of Phox2a dominates over Sim1. In fact, such a relationship between Phox2a and Sim1 activities would explain the sequential generation of OMNs and RNNs in the OMN progenitor domain, and also the premature generation of RNNs after genetic ablation of Phox2a or after the loss of Phox2a expression in Lmx1bmutants (Fig. 7). Thus, taken together, our genetic analysis indicates that the function of Lmx1a is devoted to the DN lineage, whereas Lmx1b has a more broad function and influences the specification of multiple neuronal subtypes in the ventral midbrain. In particular, Lmx1b, Phox2a and possibly Sim1 appear to be integral parts of a novel gene regulatory circuitry that controls both temporal and spatial aspects of neuronal fate determination in the developing midbrain (Fig. 7).
Acknowledgements
[image:8.612.52.252.55.171.2]We thank T. Jessell and M. Mendelsohn for providing Lmx1bmutant mice, and Jose Dias for valuable discussions. This work was supported by the Knut och Alice Wallenberg Foundation, by the EU 7th Framework Programme, by NeuroStemcell(222943) (to J.E. and T.P.), by mdDANEURODEV (to T.P.), by Vetenskapsrådet via support to the Linné Center for Developmental Biology and Regenerative Medicine, by Vetenskapsrådet (to J.E. and T.P.), by Göran Gustafssons Stiftelse, and by the Swedish Foundation for Strategic Research and research funds of the Karolinska Insititutet (to J.E.).
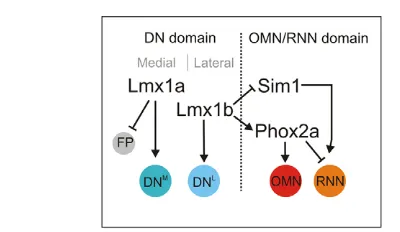
Fig. 7. Model outlining specific roles for Lmx1a, Lmx1b and Phox2a in the spatiotemporal control of neuronal fate
determination in the midbrain.Lmx1a and Lmx1b are co-expressed in DN progenitors in the ventral midbrain, and mediate both specific and redundant activities in DN specification. The DN progenitor domain can be subdivided into medial (DNM) and lateral (DNL) subpopulations of cells. There is specific requirement for Lmx1a in regulating the onset of neurogenesis in medial cells, and this process involves establishment of Notch signaling and a concomitant conversion of non-neuronal floor-plate (FP) into neuronal DN progenitors. Lmx1b has little effect on midline neurogenesis but is instead required for the establishment of lateral DN progenitors, and for suppressing Sim1 expression and RNN fate within the DN progenitor domain. This spatial patterning activity of Lmx1b is probably essential at early time points when Lmx1b is broadly expressed in the ventral midbrain but Lmx1a expression is restricted to floor-plate cells. In addition to its role in the DN differentiation lineage, Lmx1b also controls the sequential generation of OMNs and RNNs by ensuring the activation of Phox2a in progenitors located immediately lateral to DN progenitors. Phox2a and Sim1 are co-expressed within this progenitor pool, and Phox2a is required to promote OMN fate and suppress the generation of RNNs at early developmental stages. Sim1 is likely to contribute to the specification of RNNs within this
progenitor pool, but such activity must be subordinated to Phox2a and therefore not revealed until the expression of Phox2a is
downregulated.
D
E
V
E
LO
P
M
E
N
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at
http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.065482/-/DC1
References
Akazawa, C., Sasai, Y., Nakanishi, S. and Kageyama, R.(1992). Molecular characterization of a rat negative regulator with a basic Helix-Loop-Helix structure predominantly expressed in the developing nervous-system. J. Biol. Chem.267, 21879-21885.
Andersson, E., Jensen, J. B., Parmar, M., Guillemot, F. and Bjorklund, A.
(2006a). Development of the mesencephalic dopaminergic neuron system is compromised in the absence of neurogenin 2. Development133, 507-516.
Andersson, E., Tryggvason, U., Deng, Q., Friling, S., Alekseenko, Z., Robert, B., Perlmann, T. and Ericson, J.(2006b). Identification of intrinsic determinants of midbrain dopamine neurons. Cell124, 393-405.
Arenas, E.(2010). Towards stem cell replacement therapies for Parkinson’s disease. Biochem. Biophys. Res. Commun.396, 152-156.
Bjorklund, A. and Dunnett, S. B.(2007). Dopamine neuron systems in the brain: an update. Trends Neurosci.30, 194-202.
Briscoe, J., Pierani, A., Jessell, T. M. and Ericson, J.(2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell101, 435-445.
Cajanek, L., Ribeiro, D., Liste, I., Parish, C. L., Bryja, V. and Arenas, E.(2009). Wnt/beta-Catenin signaling blockade promotes neuronal induction and dopaminergic differentiation in embryonic stem cells. Stem Cells27, 2917-2927.
Chen, H., Lun, Y., Ovchinnikov, D., Kokubo, H., Oberg, K. C., Pepicelli, C. V., Gan, L., Lee, B. and Johnson, R. L.(1998). Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat. Genet. 19, 51-55.
Chizhikov, V. V. and Millen, K. J.(2004). Control of roof plate development and signaling by Lmx1b in the caudal vertebrate CNS. J. Neurosci. 24, 5694-5703.
Chung, S., Leung, A., Han, B. S., Chang, M. Y., Moon, J. I., Kim, C. H., Hong, S., Pruszak, J., Isacson, O. and Kim, K. S.(2009). Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway. Cell Stem Cell5, 646-658.
Ericson, J., Muhr, J., Jessell, T. M. and Edlund, T.(1995). Sonic hedgehog: a common signal for ventral patterning along the rostrocaudal axis of the neural tube. Int. J. Dev. Biol. 39, 809-816.
Ferri, A. L. M., Lin, W., Mavromatakis, Y. E., Wang, J. C., Sasaki, H., Whitsett, J. A. and Ang, S. L.(2007). Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development134, 2761-2769.
Fode, C., Gradwohl, G., Morin, X., Dierich, A., LeMeur, M., Goridis, C. and Guillemot, F.(1998). The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron20, 483-494.
Friling, S., Andersson, E., Thompson, L. H., Jonsson, M. E., Hebsgaard, J. B., Nanou, E., Alekseenko, Z., Marklund, U., Kjellander, S., Volakakis, N. et al.
(2009). Efficient production of mesencephalic dopamine neurons by Lmx1a expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA106, 7613-7618.
Gunhaga, L., Marklund, M., Sjodal, M., Hsieh, J. C., Jessell, T. M. and Edlund, T.(2003). Specification of dorsal telencephalic character by sequential Wnt and FGF signaling.Nat. Neurosci.6, 701-707.
Guo, C., Qiu, H. Y., Huang, Y., Chen, H., Yang, R. Q., Chen, S. D., Johnson, R. L., Chen, Z. F. and Ding, Y. Q.(2007). Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development134, 317-325.
Haber, S. N. and Fudge, J. L.(1997). The primate substantia nigra and VTA: integrative circuitry and function. Crit. Rev. Neurobiol.11, 323-342.
Hasan, K. B., Agarwala, S. and Ragsdale, C. W.(2010). PHOX2A regulation of oculomotor complex nucleogenesis. Development137, 1205-1213.
Hedlund, E. and Perlmann, T.(2009). Neuronal cell replacement in Parkinson’s disease. J. Intern. Med.266, 358-371.
Hynes, M. and Rosenthal, A.(1999). Specification of dopaminergic and serotonergic neurons in the vertebrate CNS.Curr. Opin. Neurobiol.9, 26-36.
Jacob, J., Tiveron, M. C., Brunet, J. F. and Guthrie, S.(2000). Role of the target in the pathfinding of facial visceral motor axons. Mol. Cell. Neurosci.16, 14-26.
Jessell, T. M.(2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes.Nat. Rev. Genet.1, 20-29.
Joksimovic, M., Anderegg, A., Roy, A., Campochiaro, L., Yun, B., Kittappa, R., McKay, R. and Awatramani, R.(2009a). Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proc. Natl. Acad. Sci. USA106, 19185-19190.
Joksimovic, M., Yun, B. A., Kittappa, R., Anderegg, A. M., Chang, W. W., Taketo, M. M., McKay, R. D. and Awatramani, R. B.(2009b). Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis.Nat. Neurosci.
12, 125-131.
Kala, K., Haugas, M., Lillevali, K., Guimera, J., Wurst, W., Salminen, M. and Partanen, J.(2009). Gata2 is a tissue-specific post-mitotic selector gene for midbrain GABAergic neurons. Development136, 253-262.
Kele, J., Simplicio, N., Ferri, A. L., Mira, H., Guillemot, F., Arenas, E. and Ang, S. L.(2006). Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development133, 495-505.
Kim, S. Y., Choi, K. C., Chang, M. S., Kim, M. H., Na, Y. S., Lee, J. E., Jin, B. K., Lee, B. H. and Baik, J. H.(2006). The dopamine D2 receptor regulates the development of dopaminergic neurons via extracellular signal-regulated kinase and Nurr1 activation. J. Neurosci. 26, 4567-4576.
Lek, M., Dias, J. M., Marklund, U., Uhde, C. W., Kurdija, S., Lei, Q., Sussel, L., Rubenstein, J. L., Matise, M. P., Arnold, H. H. et al.(2010). A homeodomain feedback circuit underlies step-function interpretation of a Shh morphogen gradient during ventral neural patterning. Development137, 4051-4060.
Lin, W., Metzakopian, E., Mavromatakis, Y. E., Gao, N., Balaskas, N., Sasaki, H., Briscoe, J., Whitsett, J. A., Goulding, M., Kaestner, K. H. et al.(2009). Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev. Biol. 333, 386-396.
Lindsell, C. E., Boulter, J., diSibio, G., Gossler, A. and Weinmaster, G.(1996). Expression patterns of jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol. Cell. Neurosci.8, 14-27.
Marsden, C. D.(1994). Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry57, 672-681.
McMahon, A. P. and Bradley, A.(1990). The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell62, 1073-1085.
Millonig, J. H., Millen, K. J. and Hatten, M. E.(2000). The mouse Dreher gene Lmx1a controls formation of the roof plate in the vertebrate CNS. Nature403, 764-769.
Muhr, J., Andersson, E., Persson, M., Jessell, T. M. and Ericson, J.(2001). Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell104, 861-873.
Nakano, M., Yamada, K., Fain, J., Sener, E. C., Selleck, C. J., Awad, A. H., Zwaan, J., Mullaney, P. B., Bosley, T. M. and Engle, E. C.(2001). Homozygous mutations in ARIX(PHOX2A) result in congenital fibrosis of the extraocular muscles type 2. Nat. Genet. 29, 315-320.
Nakatani, T., Minaki, Y., Kumai, M. and Ono, Y.(2007). Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development134, 2783-2793.
Nakatani, T., Kumai, M., Mizuhara, E., Minaki, Y. and Ono, Y.(2010). Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of
dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev. Biol. 339, 101-113.
Neuman, T., Keen, A., Knapik, E., Shain, D., Ross, M., Nornes, H. O. and Zuber, M. X.(1993). ME1 and GE1: basic helix-loop-helix transcription factors expressed at high levels in the developing nervous system and in
morphogenetically active regions. Eur. J. Neurosci. 5, 311-318.
Ohtsuka, T., Ishibashi, M., Gradwohl, G., Nakanishi, S., Guillemot, F. and Kageyama, R.(1999). Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J.18, 2196-2207.
Ono, Y., Nakatani, T., Sakamoto, Y., Mizuhara, E., Minaki, Y., Kumai, M., Hamaguchi, A., Nishimura, M., Inoue, Y., Hayashi, H. et al.(2007). Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development134, 3213-3225.
Pattyn, A., Morin, X., Cremer, H., Goridis, C. and Brunet, J. F.(1997). Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development124, 4065-4075.
Pattyn, A., Goridis, C. and Brunet, J. F.(2000). Specification of the central noradrenergic phenotype by the homeobox gene Phox2b. Mol. Cell. Neurosci.
15, 235-243.
Pattyn, A., Vallstedt, A., Dias, J. M., Samad, O. A., Krumlauf, R., Rijli, F. M., Brunet, J. F. and Ericson, J.(2003). Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev.17, 729-737.
Perez-Balaguer, A., Puelles, E., Wurst, W. and Martinez, S.(2009). Shh dependent and independent maintenance of basal midbrain. Mech. Dev.126, 301-313.
Pierani, A., Brenner-Morton, S., Chiang, C. and Jessell, T. M.(1999). A sonic hedgehog-independent, retinoid-activated pathway of neurogenesis in the ventral spinal cord. Cell97, 903-915.
Placzek, M. and Briscoe, J.(2005). The floor plate: multiple cells, multiple signals. Nat. Rev. Neurosci.6, 230-240.
Prakash, N., Brodski, C., Naserke, T., Puelles, E., Gogoi, R., Hall, A., Panhuysen, M., Echevarria, D., Sussel, L., Weisenhorn, D. M. et al.(2006). A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development133, 89-98.
D
Prakash, N., Puelles, E., Freude, K., Trumbach, D., Omodei, D., Di Salvio, M., Sussel, L., Ericson, J., Sander, M., Simeone, A. et al.(2009). Nkx6-1 controls the identity and fate of red nucleus and oculomotor neurons in the mouse midbrain. Development136, 2545-2555.
Puelles, E., Acampora, D., Lacroix, E., Signore, M., Annino, A., Tuorto, F., Filosa, S., Corte, G., Wurst, W., Ang, S. L. et al.(2003). Otx dose-dependent integrated control of antero-posterior and dorso-ventral patterning of midbrain. Nat. Neurosci.6, 453-460.
Sasaki, H. and Hogan, B. L.(1994). HNF-3 beta as a regulator of floor plate development. Cell76, 103-115.
Scholpp, S., Lohs, C. and Brand, M.(2003). Engrailed and Fgf8 act synergistically to maintain the boundary between diencephalon and mesencephalon. Development130, 4881-4893.
Smidt, M. P., Asbreuk, C. H., Cox, J. J., Chen, H., Johnson, R. L. and Burbach, J. P.(2000). A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b.Nat. Neurosci.3, 337-341.
Su, H., Mills, A. A., Wang, X. and Bradley, A.(2002). A targeted X-linked CMV-Cre line. Genesis32, 187-188.
Thomas, M.(2010). Role of transcription factors in cell replacement therapies for neurodegenerative conditions. Regen. Med.5, 441-450.
Van den Heuvel, D. M. and Pasterkamp, R. J.(2008). Getting connected in the dopamine system. Prog. Neurobiol.85, 75-93.
Ye, W., Shimamura, K., Rubenstein, J. L., Hynes, M. A. and Rosenthal, A.
(1998). FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell93, 755-766.