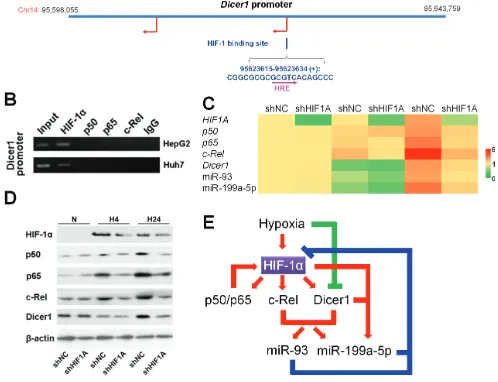
Temporal regulation of HIF-1 and NF-κB in hypoxic hepatocarcinoma cells
Full text
Figure




Related documents
In addition to our audit of the annual accounts and consolidated accounts, we have also audited the proposed appropriations of the company’s profit or loss and the administration
Molecular characterization investigates polymorphis m in selected protein molecules and DNA markers in order to evaluate genetic variation at population level as result, molecular
The study is of significance as the recommendations based on the research findings would indeed be useful to (i) The stock market policy makers, a study on the
We used a phylogenetic, comparative approach to analyse the relationship of degree of tolerance, and of FID in urban and rural populations more directly, to relative sizes of
Comparisons of modelled and measured NO 2 and PM 10 data for 1999 have been made at monitoring sites within the area and predicted future concentrations for 2004 for PM 10 and
It would appear that no work was done on the exact evaluations of multiple sums be- tween Lorenz[3] and Hardy[2] and then nothing until 1973. But the numerical evaluation of
In this study, we analyzed the quality of sequence reads and SNP detection performance using three commercially available next-generation sequencers, i.e., Roche Genome Sequencer