J
OURNAL OF
C
LINICAL
O
NCOLOGY
O R I G I N A L R E P O R TCharlotte Fribbens, Ben O’Leary, Sarah Hrebien, Isaac Garcia-Murillas, Matthew Beaney, Mitch Dowsett, and Nicholas C. Turner, Institute of Cancer Research; Charlotte Fribbens, Ben O’Leary, Stephen R.D. Johnston, and Nicholas C. Turner, Royal Marsden Hospital; Lucy Kilburn and Judith M. Bliss, The Institute of Cancer Research Clinical Trials and Statistics Unit, London, UK; Massimo Cristofanilli, Thomas Jefferson University, Philadelphia, PA; Fabrice Andre, Institut Gustave Roussy, Villejuif, France; Sherene Loi, University of Melbourne, Melbourne, Australia; Sibylle Loibl, German Breast Group, Neu-Isenburg, Germany; and John Jiang, Cynthia Huang Bartlett, and Maria Koehler, Pfizer, New York, NY.
Published online ahead of print at
www.jco.orgon June 6, 2016. Processed as a Rapid Communication manuscript.
Supported by The Royal Marsden Cancer Charity—Le Cure Fund; the Medical Research Council, Grant No. MR/ N002121/1; Breast Cancer Now with support from the Mary-Jean Mitchell Green Foundation; Cancer Research UK, Grant No. C30746/A16642; and Pfizer. The SoFEA trial was funded by Cancer Research UK, Grant No. C1491/A10962, reference Nos. CRUKE/03/021 and CRUK/ 09/007, and an educational grant from AstraZeneca. The Institute of Cancer Research Clinical Trials and Statistics Unit receives program grant funding from Cancer Research UK, Grant No. C1491/ A15955. We acknowledge National Institute for Health Research funding to the Royal Marsden and Institute of Cancer Research Biomedical Research Centre. C.F. and B.O’L. contributed equally to this work.
Authors’disclosures of potential conflicts of interest are found in the article online at
www.jco.org. Author contributions are found at the end of this article. Clinical trial information: NCT01942135, NCT00253422.
Corresponding author: Nicholas C. Turner, The Royal Marsden and Institute of Cancer Research, 237 Fulham Rd, London, SW3 6JB, UK; e-mail: nicholas.turner@icr.ac.uk.
© 2016 by American Society of Clinical Oncology
0732-183X/16/3425w-2961w/$20.00 DOI: 10.1200/JCO.2016.67.3061
Plasma
ESR1
Mutations and the Treatment of Estrogen
Receptor
–
Positive Advanced Breast Cancer
Charlotte Fribbens, Ben O’Leary, Lucy Kilburn, Sarah Hrebien, Isaac Garcia-Murillas, Matthew Beaney, Massimo Cristofanilli, Fabrice Andre, Sherene Loi, Sibylle Loibl, John Jiang, Cynthia Huang Bartlett, Maria Koehler, Mitch Dowsett, Judith M. Bliss, Stephen R.D. Johnston, and Nicholas C. Turner
See accompanying editorial on page 2950
A B S T R A C T
Purpose
ESR1mutations are selected by prior aromatase inhibitor (AI) therapy in advanced breast cancer. We assessed the impact ofESR1mutations on sensitivity to standard therapies in two phase III ran-domized trials that represent the development of the current standard therapy for estrogen receptor–positive advanced breast cancer.
Materials and Methods
In a prospective-retrospective analysis, we assessedESR1mutations in available archived baseline plasma from the SoFEA (Study of Faslodex Versus Exemestane With or Without Arimidex) trial, which compared exemestane with fulvestrant-containing regimens in patients with prior sensitivity to nonsteroidal AI and in baseline plasma from the PALOMA3 (Palbociclib Combined With Fulvestrant in Hormone Receptor–Positive HER2-Negative Metastatic Breast Cancer After Endocrine Failure) trial, which com-pared fulvestrant plus placebo with fulvestrant plus palbociclib in patients with progression after receiving prior endocrine therapy.ESR1mutations were analyzed by multiplex digital polymerase chain reaction.
Results
In SoFEA,ESR1mutations were found in 39.1% of patients (63 of 161), of whom 49.1% (27 of 55) were polyclonal, with rates of mutation detection unaffected by delays in processing of archival plasma. Patients withESR1mutations had improved progression-free survival (PFS) after taking fulvestrant (n = 45) compared with exemestane (n = 18; hazard ratio [HR], 0.52; 95% CI, 0.30 to 0.92;P= .02), whereas patients with wild-typeESR1had similar PFS after receiving either treatment (HR, 1.07; 95% CI, 0.68 to 1.67;P= .77). In PALOMA3,ESR1mutations were found in the plasma of 25.3% of patients (91 of 360), of whom 28.6% (26 of 91) were polyclonal, with mutations associated with acquired resistance to prior AI. Fulvestrant plus palbociclib improved PFS compared with fulvestrant plus placebo in bothESR1mutant (HR, 0.43; 95% CI, 0.25 to 0.74;P= .002) andESR1wild-type patients (HR, 0.49; 95% CI, 0.35 to 0.70;P,.001).
Conclusion
ESR1mutation analysis in plasma after progression after prior AI therapy may help direct choice of further endocrine-based therapy. Additional confirmatory studies are required.
J Clin Oncol 34:2961-2968. © 2016 by American Society of Clinical Oncology
INTRODUCTION
Targeting the estrogen receptor (ER) with endocrine
therapies was thefirst molecularly targeted treatment
of breast cancer and remains a mainstay of treatment
of all stages of ER-positive disease.1-3Approximately
75% of breast cancers are ER-positive, with endo-crine therapy the favored initial choice for patients
who develop metastatic disease.4In this setting,
al-most all patients will acquire endocrine resistance,
with a proportion demonstrating primary re-sistance. Identifying therapies with activity in tumors resistant to standard endocrine therapy is a key therapeutic challenge.
Although diverse mechanisms of resistance to endocrine therapy have been described, recent
evidence has identified mutations in the ER gene
(ESR1).5ESR1mutations occur rarely in primary
breast cancer,6 but have a high prevalence in
advanced breast cancers previously treated with
through selective treatment pressure. MostESR1mutations occur in hotspot regions in the ligand-binding domain of ER, resulting in
ligand-independent, constitutive ER activity.8-11Prior research has
demonstrated that circulating tumor DNA (ctDNA) is detected in the plasma of patients with cancer and may provide a robust,
noninvasive method for detectingESR1mutations.7,12-15
The most effective treatment ofESR1mutant breast cancer is
uncertain.8,9 In a retrospective, single-center analysis, we have
demonstrated resistance to subsequent AI-based therapy in
pa-tients withESR1 mutations in plasma.7 Preclinical studies have
reported growth inhibition of ESR1 mutant cell lines with
fulvestrant, a selective ER degrader, but with less sensitivity to fulvestrant than wild-type ER, and there is uncertainty whether the
required doses are achieved clinically.8 Palbociclib, a CDK4/6
inhibitor, has demonstrated substantial clinical activity in
com-bination with both fulvestrant and AIs.16,17 CDK4/6 inhibition
resensitizes cells with in vitro-derived resistance to endocrine
therapy,18andESR1mutant models are sensitive to combinations
of selective ER degraders with palbociclib.19
In this study, we used ctDNA analysis to identifyESR1mutant
cancers and assess the impact of mutations on the efficacy of
current therapies. Baseline plasma samples were drawn from two
SoFEA cohort (N = 723)
Fulvestrant containing (n = 474)
Available plasma (n = 105)
Mutation analyzed (n = 104)
ESR1 mutant (n = 45)
Unable to analyze (n = 1) No available plasma
(n = 369)
ESR1 wild type (n = 59)
Exemestane (n = 249)
Available plasma (n = 57)
Mutation analyzed (n = 57)
ESR1 mutant (n = 18)
ESR1 wild type (n = 39)
* * No available plasma(n = 192)
Unable to analyze (n = 0)
B
PALOMA3 cohort (N = 521)
Fulvestrant and placebo (n = 174)
Available plasma (n = 120)
Mutation analyzed (n = 120)
ESR1 mutant (n = 28)
Unable to analyze (n = 0) No available plasma
(n = 54)
ESR1 wild type (n = 92)
Fulvestrant and palbociclib
(n = 347)
Available plasma (n = 240)
Mutation analyzed (n = 240)
ESR1 mutant (n = 63)
ESR1 wild type (n = 177) No available plasma
(n = 107)
Unable to analyze (n = 0)
A
randomized phase III studies that spanned the development of standard endocrine-based therapy for breast cancer progressing
after prior endocrine therapy.17,20The SoFEA (Study of Faslodex
Versus Exemestane With or Without Arimidex) trial showed no
significant difference in its primary end point of progression free
survival (PFS) between fulvestrant 250 mg, fulvestrant 250 mg plus anastrozole, and exemestane in a population previously sensitive
to AIs.20The PALOMA3 (Palbociclib Combined With Fulvestrant
in Hormone Receptor–Positive HER2-Negative Metastatic Breast
Cancer After Endocrine Failure) study demonstrated that palbo-ciclib improves PFS (its primary end point) when added to ful-vestrant 500 mg in patients with progression after receiving prior
endocrine therapy.17 From our prior retrospective study,7we
hy-pothesized thatESR1mutant patients would have a poor prognosis
when treated with exemestane and that prognosis would be im-proved with fulvestrant (samples from SoFEA), with additional improvement with palbociclib (samples from PALOMA3).
MATERIALS AND METHODS
The SoFEA study was a multicenter, randomized phase III trial in post-menopausal women with advanced, hormone receptor–positive breast cancer who had demonstrated prior sensitivity to AIs. Sensitivity was defined as relapse or progression after taking a nonsteroidal AI given as adjuvant treatment for at least 12 months or as first-line metastatic treatment for at least 6 months.20 Patients were assigned fulvestrant
(500 mg intramuscularly on day 1, followed by 250 mg on days 15 and 29, then every 28 days) plus anastrozole 1 mg, fulvestrant plus placebo, or exemestane 25 mg. Baseline plasma samples were available from 162 patients of the 723 enrolled (22.4%), with no samples available before January 2, 2008, because of afire at the Royal Marsden Hospital (Fig 1A). The subset of patients with baseline plasma samples available had similar characteristics, except for a longer time to relapse and a longer time taking an AI, and outcomes similar to the rest of the study population (Data Supplement). Written informed consent was obtained from all participants, and ESR1analysis was approved by the research ethics committee.
The PALOMA3 trial was a multicenter, randomized phase III trial assessing palbociclib and fulvestrant in premenopausal and post-menopausal women with advanced, hormone receptor–positive breast cancer who had progressed during prior endocrine therapy.17Patients were
assigned 2:1 to palbociclib (125 mg orally for 3 weeks followed by 1 week off) and fulvestrant (500 mg intramuscularly every 14 days for thefirst three injections, then 500 mg every 28 days), or matching placebo plus fulvestrant. Premenopausal patients received goserelin for the study du-ration. We analyzed 360 baseline plasma samples from 521 patients (69.1%) enrolled in the PALOMA3 trial (Fig 1B). The subset of patients with baseline plasma samples available had similar characteristics, with the exception of prior chemotherapy exposure, and outcomes similar to the rest of the study population (Data Supplement). Written informed consent was obtained from all participants.
Processing of Plasma and Extraction of Circulating DNA
In the SoFEA trial, baseline blood was collected in EDTA blood collection tubes and processed within 0 to 9 days of sample collection. Plasma was separated by centrifugation at 1,600 g for 20 minutes. In the PALOMA3 study, baseline blood was collected in EDTA tubes and centrifuged within 30 minutes at 1,500 to 2,000 g for 10 minutes. Samples were stored at280°C until DNA extraction. DNA extraction was per-formed using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Digital Polymerase Chain Reaction Analysis
Total free DNA was quantified from plasma using RNase P as the ref-erence gene as previously reported7(Data Supplement). ForESR1mutation analysis, we used commercially available multiplex droplet digital polymer-ase chain reaction (ddPCR) assays for the seven most commonESR1 mu-tations; multiplex 1 included c.1138G.C(E380Q), c.1607T.G(L536R), c.1610A.G(Y537C), and c.1613A.G(D538G; dHsaMDXE91450042); and multiplex 2 included c.1387T.C(S463P), c.1609T.A(Y537N), and c.1610A.C(Y537S; dHsaMDXE65719815). ddPCR was performed on a QX200 system (Bio-Rad, Hercules, CA) on a minimum of 1-mL plasma equivalent for SoFEA samples and 0.5-mL for PALOMA3. A multiplex assay was considered mutation positive if at least twoESR1mutant droplets were observed. The results obtained on the multiplex ddPCR were further characterized using uniplex ddPCR assays. Mutation allele fraction and copies per mL were calculated as previously described.21A sample was considered polyclonal if it was positive on both multiplexes or if separate mutations were characterized on uniplex confirmation.
Validation of Analysis ofESR1Mutations in Archival Plasma Samples
Archival plasma samples were available in the SoFEA trial, and we validated the analysis of ctDNA in archival plasma, demonstrating that archival EDTA plasma samples can be used for ctDNA analysis with ddPCR (Data Supplement).
Statistical Analysis
In this prospective-retrospective study, the most recent clinical data snapshots were used for both SoFEA and PALOMA3.20,22ESR1mutation
status was measured as a binary outcome (mutated v wild type). The principal analysis population for both trials included all patients who were randomly assigned on an intention-to-treat basis and had been assessed for
ESR1status. There was no difference in efficacy between the two fulvestrant groups in the SoFEA trial, and these were merged for primary end point analysis, as prespecified in the statistical analysis plan.Pvalues were two tailed and considered significant ifPwas,.05 for the principal analyses of PFS. Other analyses were exploratory and considered significant ifPwas
,.01 to take multiple comparisons into account.
Survival end point comparisons were made using the log-rank test. Hazard ratios (HRs) were obtained from Cox proportional hazards re-gression models. The proportionality assumption of the Cox models was tested with Schoenfeld residuals and was shown to hold for all analyses. Survival data in PALOMA3 were stratified according to the presence or absence of visceral disease and sensitivity to prior endocrine therapy in line with the main trial analyses. Interaction tests were used to explore dif-ferential effects betweenESR1mutation status and trial treatment in re-lation to PFS. Multivariable models assessed ESR1mutation status and treatment group separately for each trial, adjusting for, in the case of SoFEA, factors identified for the principal analysis, namely, time from diagnosis tofirst relapse, number of disease sites present at baseline, and prior AI setting and time receiving an AI. Any baseline characteristics that were statistically significant when comparingESR1mutation versus wild type were also included in the multivariable models separately for SoFEA and PALOMA3 if they significantly added value to the model (likelihood ratio test P ,.05). All statistical analyses were performed with Stata (version 13.1; STATA, College Station, TX) or GraphPad Prism (version 6.0; GraphPad Software, La Jolla, CA).
RESULTS
Impact ofESR1Mutation on Sensitivity to Endocrine
Therapies in SoFEA
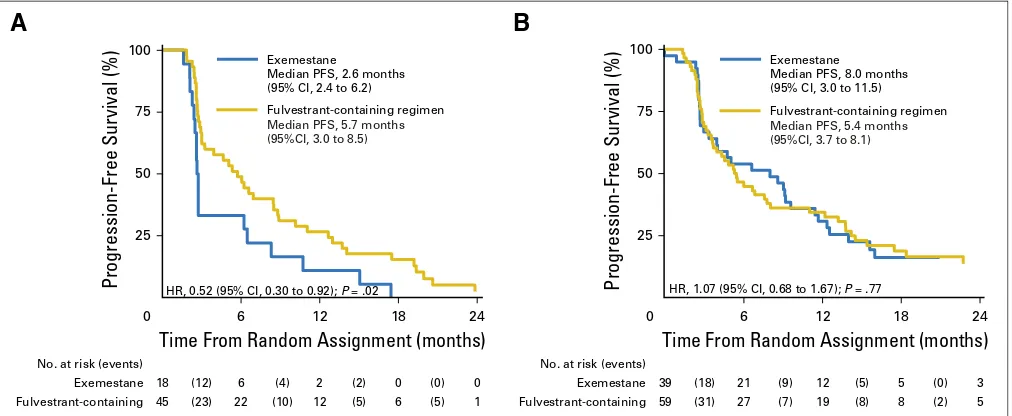
In SoFEA,ESR1mutation status was successfully analyzed in
baseline plasma samples, withESR1mutations detected in 39.1%
of samples (63 of 161). We assessed the impact ofESR1mutations
on outcome in patients randomly assigned to receive exemestane (n = 57) versus fulvestrant-containing (n = 104) regimens. For
patients withESR1mutant ctDNA, the median PFS was 2.6 months
(95% CI, 2.4 to 6.2) for patients given exemestane and 5.7 months
(95% CI, 3.0 to 8.5) for those given fulvestrant (Fig 2A; HR, 0.52;
95% CI, 0.30 to 0.92;P= .02), whereas patients with wild-type
ESR1had a median PFS of 8.0 months (95% CI, 3.0 to 11.5) when
given exemestane and a median PFS of 5.4 months (95% CI, 3.7 to
8.1) when given fulvestrant (Fig 2B; HR, 1.07; 95% CI, 0.68 to 1.67;
P= .77). The interaction test between treatment allocation and
ESR1mutation status was P= .07. Considering ESR1mutation
status within the exemestane group, patients with anESR1
mu-tation had worse PFS thanESR1wild type (Data Supplement; HR,
2.12; 95% CI, 1.18 to 3.81; P = .01). In the SoFEA study, the
number of deaths provided insufficient statistical power to detect
a statistically significant difference in survival curves, although the
effects ofESR1mutation on overall survival in patients treated
with exemestane were consistent with the PFS analysis (Data Supplement).
Impact ofESR1Mutation on Sensitivity to Palbociclib in
PALOMA3
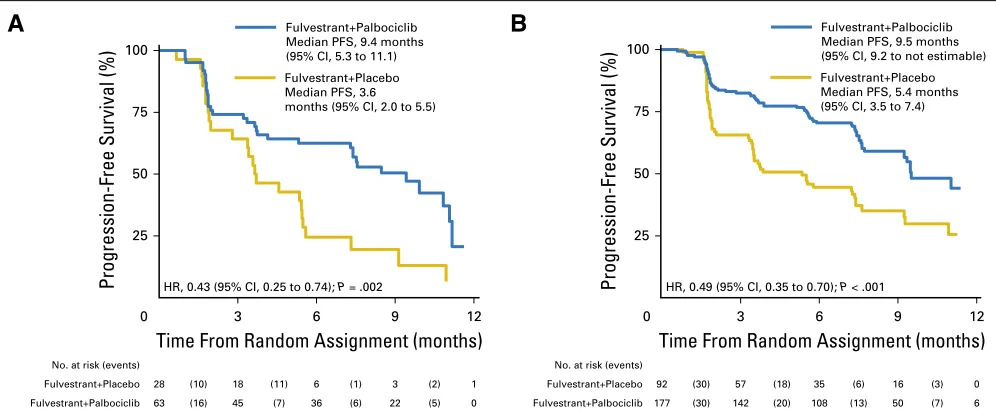
In PALOMA3,ESR1mutation status was successfully analyzed
in 100% of available samples (360 of 360, representing 69.1% of all
patients), withESR1mutations detected in 25.3% of patients (91 of
360). For patients withESR1mutant ctDNA, the median PFS was
9.4 months (95% CI, 5.3 to 11.1) for those taking fulvestrant and palbociclib, compared with 3.6 months (95% CI, 2.0 to 5.5) for
those taking fulvestrant and placebo (Fig 3A; HR, 0.43; 95% CI,
0.25 to 0.74;P= .002). ForESR1wild-type patients, the PFS was
9.5 months (95% CI, 9.2 to not estimable) for those taking ful-vestrant and palbociclib and 5.4 months (95% CI, 3.5 to 7.4) for
those taking fulvestrant and placebo (Fig 3B; HR, 0.49; 95% CI,
0.35 to 0.70;P,.001). The benefit from palbociclib was therefore
seen despite ESR1 mutation status (interaction P = .74). The
confirmed objective response rates were not significantly different
between the ESR1 mutant and ESR1 wild-type patients (Data
Supplement), but a negative impact ofESR1mutations on response
to fulvestrant and palbociclib cannot be excluded.
Clinical and Pathologic Associations ofESR1Mutations
With different ESR1 mutation rates observed between the
studies, we investigated which clinical and pathologic features were
associated with ESR1mutations. SoFEA recruited a relatively
ho-mogenous population of postmenopausal women with prior
sen-sitivity to an AI, and there were no significant differences in baseline
characteristics between patients with and withoutESR1mutations
(Data Supplement).
A more diverse population of patients whose disease had progressed after receiving prior endocrine therapy was recruited
for PALOMA3. As in a prior retrospective study,7ESR1mutations
were almost exclusively found in patients with prior AI exposure with or without tamoxifen and were rare in patients with prior
tamoxifen exposure only (Table 1; 28.9% [90 of 311]v2.0% [one
of 49], respectively;P,.001).ESR1mutation was associated with
sensitivity to prior endocrine therapy (sensitive to prior endocrine
therapy, 29.8% [85 of 285]vresistant, 8.0% [six of 75];P,.001).
ESR1mutation was significantly associated with bone metastases
(P= .001) and prior lines of therapy for metastatic disease (P= .01;
Table 1). In multivariable analysis,ESR1mutation status remained
significantly associated with exposure to an AI and sensitivity to
endocrine therapy, and with bone or visceral disease (Data Supplement).
Impact of Individual Mutations and Clonality
In PALOMA3 overall, patients with ESR1 mutations had
marginal statistical significance toward worse PFS compared with
ESR1wild type in both univariate analysis (HR, 1.46; 95% CI, 1.06
Progression-Free Survival (%)
100
75
HR, 0.52 (95% CI, 0.30 to 0.92); P = .02
50
25
No. at risk (events)
18 (12) 6 (4) 2 (2) 0 (0) 0 Exemestane
(23) (10) (5) (5)
45 22 12 6 1
Fulvestrant-containing
Time From Random Assignment (months)
0 6 12 18 24
A
HR, 1.07 (95% CI, 0.68 to 1.67); P = .77
Progression-Free Survival (%)
100
75
50
25
No. at risk (events)
39 (18) 21 (9) 12 (5) 5 (0) 3 Exemestane
(31) (7) (8) (2)
59 27 19 8 5
Fulvestrant-containing
Time From Random Assignment (months)
0 6 12 18 24
B
Exemestane
Median PFS, 2.6 months (95% CI, 2.4 to 6.2)
Fulvestrant-containing regimen
Median PFS, 5.7 months (95%CI, 3.0 to 8.5)
Exemestane
Median PFS, 8.0 months (95% CI, 3.0 to 11.5)
Fulvestrant-containing regimen
Median PFS, 5.4 months (95%CI, 3.7 to 8.1)
to 2.02; P = .02; Data Supplement) and multivariable analysis
(HR, 1.49; 95% CI, 1.07 to 2.08;P= .02; Data Supplement). In
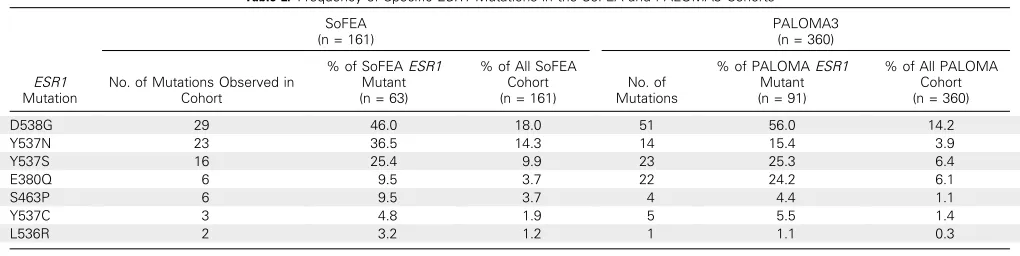
both studies, there was a predominance of mutations in D538G,
Y537N, Y537S, and E380Q (Table 2). Mutations were polyclonal
in 49.1% ofESR1 mutant samples (27 of 55) in SoFEA and in
28.6% (26 of 91) in PALOMA 3 (Data Supplement). The lower
rate of mutations and polyclonality in PALOMA3 likely reflects
the inclusion of patients with tamoxifen exposure and disease with intrinsic resistance to prior endocrine therapy. In vitro,
differentESR1mutations show varied sensitivity to fulvestrant,10
and we explored the impact of individual mutations on outcome with fulvestrant using a post hoc combined analysis of fulvestrant groups in both studies (fulvestrant-containing in SoFEA and
fulvestrant plus placebo in PALOMA3, n = 224). No significant
difference was observed in PFS for the individual mutations D538G, E380Q, or Y537S, or for patients with polyclonal versus
monoclonal mutations (allP..1; Data Supplement), although
these analyses are limited by their exploratory nature and sample size.
DISCUSSION
Results from this prospective-retrospective study on archival samples demonstrate that plasma DNA analysis has potential clinical utility for patients with advanced ER-positive breast cancer that has progressed after prior AI therapy. In patients from the
SoFEA trial, the detection of ESR1 mutations in plasma DNA
predicted relative resistance to exemestane and relative sensitivity
to fulvestrant. In contrast, patients without ESR1mutations
de-tected may derive further benefit from exemestane, as well as
fulvestrant. Patients with ESR1 mutant cancers have a poor
prognosis,23and the combination of palbociclib and fulvestrant
500 mg appeared to be equally effective in patients with or without
ESR1mutations (interaction testP= .74), although further studies
are required to confirm the efficacy of CDK4/6 inhibitors inESR1
mutant cancer.
Our results demonstrate that archival plasma samples collected in EDTA with substantially delayed processing can be used for ctDNA analysis using digital polymerase chain reaction (PCR). This technique is robust for mutation detection, despite the release of contaminating free germline DNA from white blood cell lysis, allowing for accurate ctDNA analysis in what are
traditionally seen as suboptimally processed samples.24 This
finding will open up large archival plasma sets linked to phase III
trials for ctDNA analysis using digital PCR. Samples were col-lected in EDTA tubes, which chelate calcium and inhibit blood
DNases,25 and it is unknown whether the findings apply to
samples collected with alternative anticoagulants or other
methods ofESR1detection. Thefinding on multivariable analysis
that detection of ESR1 mutations is associated with bone and
visceral disease may suggest limited detection in patients with nodal or locoregional recurrence only, and this should be con-sidered in future studies.
ESR1mutations are a rare cause of intrinsic primary
endo-crine resistance and are observed in advanced ER-positive breast cancer during the development of acquired secondary resistance to
AI therapy (Table 1). This is consistent with the higher rate ofESR1
mutations observed in SoFEA, where recruited patients had all
demonstrated previous clinical benefit from an AI. As a
conse-quence, there were no clinical-pathologic associations of ESR1
mutations in the SoFEA cohort (Data Supplement) that could
confound the observed predictive effects (Fig 2). The prior
hor-mone sensitivity of patients in SoFEA may contribute to the
re-sidual sensitivity ofESR1wild-type cancers to exemestane (Fig 2B;
Data Supplement), suggesting that exemestane may be still active in tumors that have acquired resistance to nonsteroidal AIs without
selection of anESR1mutation.
Progression-Free Survival (%)
100
75
HR, 0.43 (95% CI, 0.25 to 0.74); P = .002
50
25
No. at risk (events)
28 (10) 18 (11) 6 (1) 3 (2) 1
Fulvestrant+Placebo
(16) (7) (6) (5)
63 45 36 22 0
Fulvestrant+Palbociclib
Time From Random Assignment (months)
0 3 6 9 12
A
HR, 0.49 (95% CI, 0.35 to 0.70); P < .001
Progression-Free Survival (%)
100
75
50
25
No. at risk (events)
92 (30) 57 (18) 35 (6) 16 (3) 0
Fulvestrant+Placebo
(30) (20) (13) (7)
177 142 108 50 6
Fulvestrant+Palbociclib
Time From Random Assignment (months)
0 3 6 9 12
B
Fulvestrant+Palbociclib Median PFS, 9.4 months (95% CI, 5.3 to 11.1) Fulvestrant+Placebo Median PFS, 3.6 months (95% CI, 2.0 to 5.5)
Fulvestrant+Palbociclib Median PFS, 9.5 months (95% CI, 9.2 to not estimable) Fulvestrant+Placebo Median PFS, 5.4 months (95% CI, 3.5 to 7.4)
These results are consistent with our prior retrospective analysis that showed, in a single-center retrospective series, that
patients with ESR1 mutations had a poor PFS on subsequent
AI-based therapy.7Here, in a prospective-retrospective analysis
of SoFEA, we observed that patients with ESR1mutations
de-tected in plasma had poor PFS on further AI therapy, specifically,
exemestane, but relatively improved PFS when treated with
fulvestrant. This provides the first evidence of potential
clini-cal utility for the use ofESR1plasma DNA analysis in selecting
the most appropriate endocrine therapy.26 It should be noted
that although we assessed seven differentESR1mutations, there
may be other mutations or aberrations in ESR1, such as
am-plification or rearrangement, that could also contribute to AI
resistance.27
Our results suggest thatESR1mutant cancers show selective
sensitivity to fulvestrant, a drug that degrades the ER, but overall with modestly worse PFS than wild-type cancers. This is consistent
with the finding that in vitro hotspot mutations in the
ligand-binding domain partially inhibit fulvestrant ligand-binding.10More
po-tent receptor degraders may have the popo-tential to further improve
with fulvestrant inESR1mutant cancers, and a number of such
therapies are in early clinical development. Our data confirm
laboratoryfindings thatESR1mutant cancers continue to drive cell
cycle progression through cyclin D1 activation of CDK4/6 and that CDK4/6 inhibition remains a highly active therapeutic approach in
ESR1mutant cancer when combined with fulvestrant that at least
partially blocks mutant ER function.
Our study has a number of important limitations. The bi-ologic analysis was retrospective for both studies, although to
mitigate bias, prespecified statistical analysis plans were developed.
A relatively small number of baseline samples were available from the SoFEA trial (162), which limits the statistical power to detect important interactions and differences, such as the interaction test
betweenESR1mutation and relative sensitivity to fulvestrant over
Table 1.Baseline Characteristics ofESR1Mutant Patients VersusESR1Wild-Type Patients in PALOMA3
ESR1Mutant (n = 91)
ESR1Wild Type
(n = 269) P
Median age at random assignment, years (IQR) 59 (50, 66) 56 (48, 65) .2
Hormone receptor status, No. (%)* .016
ER-positive/PR-positive 69 (75.8) 173 (64.3)
ER-positive/PR-negative 17 (18.7) 87 (32.3)
Disease-free interval (months), No. (%)† .22
#24 6 (11.3) 38 (19.5)
.24 47 (88.7) 157 (80.5)
Menopausal status, No. (%) .07
Premenopausal/perimenopausal 12 (13.2) 61 (22.7)
Postmenopausal 79 (86.8) 208 (77.3)
Sensitivity to prior endocrine treatment, No. (%) ,.001
Yes 85 (93.4) 200 (74.4)
No 6 (6.6) 69 (25.7)
Visceral metastases, No. (%) .11
Yes 62 (68.1) 157 (58.4)
No 29 (31.9) 112 (41.6)
Bone metastases, No. (%) .001
Yes 80 (87.9) 191 (71.0)
No 11 (12.1) 79 (29.0)
Soft tissue/nodal metastases, No. (%) .04
Yes 28 (30.8) 118 (43.9)
No 63 (69.2) 151 (56.1)
Prior endocrine therapies, No. (%) ,.001
Tamoxifen only 1 (1.1) 48 (17.8)
AI only 41 (45.1) 103 (38.3)
AI and tamoxifen 49 (53.9) 118 (43.9)
Prior chemotherapy, No. (%) .05
Neoadjuvant/adjuvant 32 (35.2) 123 (45.7)
Metastatic6adjuvant 24 (26.4) 79 (29.4)
None 35 (38.5) 67 (24.9)
Prior lines of therapy for metastatic disease, No. (%) .01
0 14 (15.4) 67 (24.9)
1 41 (45.1) 122 (45.4)
2 22 (24.2) 63 (23.4)
3+ 14 (15.4) 17 (6.3)
Disease sites, No. (%) .74
1 32 (35.2) 81 (3.1)
2 21 (23.1) 80 (29.7)
3+ 38 (41.8) 108 (40.2)
NOTE. To correct for multiple comparisons, associations with baseline characteristics were considered significant atP,.01. Abbreviations: AI, aromatase inhibitor; ER, estrogen receptor; IQR, interquartile range; PR, progesterone receptor.
*Local testing, analysis omitsfiveESR1mutant patients and nineESR1wild-type patients classified as either ER-negative/PR-positive or ER/PR unknown.
exemestane (P= .07). Further confirmatory studies are required
before it could be concluded that plasmaESR1mutation analysis
may be used to guide treatment. Although samples were collected in multiple sites, analysis was centralized in a single laboratory. No assessment of interlaboratory agreement has yet been conducted
with the assays, and this would be required beforeESR1digital PCR
can be used in clinical decision making. The SoFEA trial recruited only patients with sensitivity to prior AIs, and it is unknown whether the results would also apply to patients who were not sensitive to prior AIs. The exploratory analyses in this report are hypothesis
generating and will require confirmation in future studies.
ESR1mutations are found at high frequency in patients who
progress after taking prior AIs and can be analyzed relatively simply
and cheaply with digital PCR. Our data suggest thatESR1mutation
analysis may have clinical utility in directing further endocrine
therapy, although further confirmatory studies are required. Our
results demonstrate thatESR1mutant and wild-type cancers seem
to be distinct subtypes of breast cancer that differ in response to standard endocrine therapies. Future clinical trials in advanced breast cancer might consider using plasma DNA analysis to optimize
endocrine therapy choice according toESR1mutation status.
AUTHORS’DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Disclosures provided by the authors are available with this article at www.jco.org.
AUTHOR CONTRIBUTIONS
Conception and design:Charlotte Fribbens, Ben O’Leary, Sherene Loi, Sibylle Loibl, John Jiang, Cynthia Huang Bartlett, Maria Koehler, Mitch Dowsett, Judith M. Bliss, Stephen R.D. Johnston, Nicholas C. Turner
Collection and assembly of data:Charlotte Fribbens, Ben O’Leary, Sarah Hrebien, Isaac Garcia-Murillas, Matthew Beaney, Sibylle Loibl, John Jiang, Cynthia Huang Bartlett, Maria Koehler, Mitch Dowsett
Data analysis and interpretation:Charlotte Fribbens, Ben O’Leary, Lucy Kilburn, Sarah Hrebien, Isaac Garcia-Murillas, Massimo Cristofanilli, Fabrice Andre, Sherene Loi, Sibylle Loibl, John Jiang, Cynthia Huang Bartlett, Maria Koehler, Mitch Dowsett, Judith M. Bliss, Nicholas C. Turner
Manuscript writing:All authors
Final approval of manuscript:All authors
Accountable for all aspects of the work:All authors
REFERENCES
1. Early Breast Cancer Trialists’Collaborative Group (EBCTCG): Effects of chemotherapy and hormonal therapy for early breast cancer on re-currence and 15-year survival: An overview of the randomised trials. Lancet 365:1687-1717, 2005
2. Smith IE, Dowsett M: Aromatase inhibitors in breast cancer. N Engl J Med 348:2431-2442, 2003
3. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG), Dowsett M, Forbes JF, et al: Aromatase inhibitors versus tamoxifen in early breast cancer: Patient-level meta-analysis of the rando-mised trials. Lancet 386:1341-1352, 2015
4. National Comprehensive Cancer Network: National Comprehensive Cancer Network Guide-lines for Breast Cancer.https://www.nccn.org/ professionals/physician_gls/f_guidelines.asp
5. Jeselsohn R, Buchwalter G, De Angelis C, et al: ESR1 mutations—A mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol 12:573-583, 2015
6. Cancer Genome Atlas Network: Comprehen-sive molecular portraits of human breast tumours. Nature 490:61-70, 2012
7. Schiavon G, Hrebien S, Garcia-Murillas I, et al: Analysis of ESR1 mutation in circulating tu-mor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 7: 313ra182, 2015
8. Jeselsohn R, Yelensky R, Buchwalter G, et al: Emergence of constitutively active estrogen receptor-amutations in pretreated advanced estro-gen receptor-positive breast cancer. Clin Cancer Res 20:1757-1767, 2014
9. Robinson DR, Wu Y-M, Vats P, et al: Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 45:1446-1451, 2013
10. Toy W, Shen Y, Won H, et al: ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 45:1439-1445, 2013
11. Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, et al: D538G mutation in estrogen receptor-a: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res 73:6856-6864, 2013
12. Siravegna G, Mussolin B, Buscarino M, et al: Clonal evolution and resistance to EGFR blockade in
the blood of colorectal cancer patients. Nat Med 21: 795-801, 2015
13. Bettegowda C, Sausen M, Leary RJ, et al: Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6: 224ra24, 2014
14. Forshew T, Murtaza M, Parkinson C, et al: Noninvasive identification and monitoring of cancer mutations by targeted deep sequenc-ing of plasma DNA. Sci Transl Med 4:136ra68, 2012
15. Dawson S-J, Tsui DWY, Murtaza M, et al: Analysis of circulating tumor DNA to monitor meta-static breast cancer. N Engl J Med 368:1199-1209, 2013
16. Finn RS, Crown JP, Lang I, et al: The cyclin-dependent kinase 4/6 inhibitor palbociclib in combi-nation with letrozole versus letrozole alone asfi rst-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol 16: 25-35, 2015
17. Turner NC, Ro J, Andr ´e F, et al: Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med 373:209-219, 2015
Table 2. Frequency of SpecificESR1Mutations in the SoFEA and PALOMA3 Cohorts
ESR1
Mutation
SoFEA (n = 161)
PALOMA3 (n = 360)
No. of Mutations Observed in Cohort
% of SoFEAESR1
Mutant (n = 63)
% of All SoFEA Cohort (n = 161)
No. of Mutations
% of PALOMAESR1
Mutant (n = 91)
% of All PALOMA Cohort (n = 360)
D538G 29 46.0 18.0 51 56.0 14.2
Y537N 23 36.5 14.3 14 15.4 3.9
Y537S 16 25.4 9.9 23 25.3 6.4
E380Q 6 9.5 3.7 22 24.2 6.1
S463P 6 9.5 3.7 4 4.4 1.1
Y537C 3 4.8 1.9 5 5.5 1.4
18. Finn RS, Dering J, Conklin D, et al: PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estro-gen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 11:R77, 2009
19. Wardell SE, Ellis MJ, Alley HM, et al: Efficacy of SERD/SERM hybrid-CDK4/6 inhibitor combina-tions in models of endocrine therapy-resistant breast cancer. Clin Cancer Res 21:5121-5130, 2015
20. Johnston SRD, Kilburn LS, Ellis P, et al: Fulvestrant plus anastrozole or placebo versus exemestane alone after progression on non-steroidal aromatase inhibitors in postmenopausal patients with hormone-receptor-positive locally advanced or metastatic breast cancer (SoFEA): A composite, multicentre, phase 3 randomised trial. Lancet Oncol 14:989-998, 2013
21. Garcia-Murillas I, Schiavon G, Weigelt B, et al: Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 7: 302ra133, 2015
22. Cristofanilli M, Turner NC, Bondarenko I, et al: Fulvestrant plus palbociclib versus fulvestrant plus pla-cebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): Final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol 17: 425-439, 2016
23. Chandarlapaty S SP, Chen D, He W, et al: [S2-07] cfDNA analysis from BOLERO-2 plasma samples identifies a high rate of ESR1 mutations: Exploratory analysis for prognostic and predictive correlation of mutations reveals different efficacy outcomes of endocrine therapy–based regimens. Presented at the
San Antonio Breast Cancer Symposium, San Anto-nio, TX, December 8-12, 2015
24. Norton SE, Lechner JM, Williams T, et al: A stabilizing reagent prevents cell-free DNA contami-nation by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem 46:1561-1565, 2013
25. Barra GB, Santa Rita TH, de Almeida Vasques J, et al: EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin Biochem 48:976-981, 2015
26. Simon RM, Paik S, Hayes DF: Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 101:1446-1452, 2009 27. Ma CX, Reinert T, Chmielewska I, et al: Mechanisms of aromatase inhibitor resistance. Nat Rev Cancer 15:261-275, 2015
AUTHORS’DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
PlasmaESR1Mutations and the Treatment of Estrogen Receptor–Positive Advanced Breast Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer towww.asco.org/rwcorjco.ascopubs.org/site/ifc.
Charlotte Fribbens
No relationship to disclose
Ben O’Leary
Research Funding:Pfizer
Lucy Kilburn
No relationship to disclose
Sarah Hrebien
No relationship to disclose
Isaac Garcia-Murillas
No relationship to disclose
Matthew Beaney
Research Funding: Randox Laboratories (I)
Massimo Cristofanilli Honoraria:Agendia
Company: Dompe Farmaceutici, Celgene, Pfizer
Consulting or Advisory Role:Dompe Farmaceutici, Cynvenio Biosystems, Newomics
Fabrice Andre
Research Funding:AstraZeneca (Inst), Novartis (Inst), Pfizer (Inst), Eli Lilly (Inst)
Sherene Loi
Research Funding:Genentech (Inst), Pfizer (Inst), Novartis (Inst), Merck (Inst)
Patents, Royalties, Other Intellectual Property:PI3K pathway gene signature granted by the European and US patent offices
Sibylle Loibl
Research Funding:Novartis (Inst), Pfizer (Inst) Genentech (Inst)
John Jiang Employment:Pfizer
Stock or Other Ownership:Pfizer
Cynthia Huang Bartlett Employment:Pfizer
Stock or Other Ownership:Pfizer
Maria Koehler Employment:Pfizer
Stock or Other Ownership:Pfizer
Mitch Dowsett
Consulting or Advisory Role:Radius
Speakers’Bureau:AstraZeneca, Myriad Genetics
Research Funding:AstraZeneca (Inst)
Judith M. Bliss
Research Funding:AstraZeneca (Inst), Pfizer (Inst), Janssen Cilag (Inst), Novartis (Inst), Roche (Inst), Clovis Oncology (Inst)
Stephen R.D. Johnston
Consulting or Advisory Role:Eli Lilly, AstraZeneca, Novartis,
Speakers’Bureau:GlaxoSmithKline, Roche
Research Funding:Pfizer (Inst)
Nicholas C. Turner
Consulting or Advisory Role:Roche, Pfizer, Novartis, AstraZeneca
Acknowledgment