Bis{4-[(
E
)-2-(1
H
-indol-3-yl)ethenyl]-1-methylpyridinium}
4-chlorobenzene-sulfonate nitrate
Hoong-Kun Fun,a*‡ Ching Kheng Quah,aNawong Boonnakband Suchada Chantraprommac§
aX-ray Crystallography Unit, School of Physics, Universiti Sains Malaysia, 11800
USM, Penang, Malaysia,bFaculty of Traditional Thai Medicine, Prince of Songkla
University, Hat-Yai, Songkhla 90112, Thailand, andcDepartment of Chemistry,
Faculty of Science, Prince of Songkla University, Hat-Yai, Songkhla 90112, Thailand Correspondence e-mail: hkfun@usm.my
Received 2 November 2013; accepted 2 November 2013
Key indicators: single-crystal X-ray study;T= 153 K; mean(C–C) = 0.005 A˚; disorder in main residue;Rfactor = 0.074;wRfactor = 0.254; data-to-parameter ratio = 9.8.
In the title mixed salt, 2C16H15N2+C6H4ClO3SNO3, one of the cations shows whole molecule disorder over two sets of sites in a 0.711 (7):0.289 (7) ratio. The 4-chorobenzenesulfon-ate anion is also disordered over two orientations in a 0.503 (6):0.497 (6) ratio. The cations are close to planar, the dihedral angles between the pyridinium and indole rings being 1.48 (3) in the ordered cation, and 5.62 (3) and 2.45 (3), respectively, for the major and minor components of the disordered cation. In the crystal, the cations are stacked in an antiparallel manner approximately along thea-axis direction and linked with the anionsviaN—H O hydrogen bonds and C—H O interactions, generating a three-dimensional
network. Weak C—H and – interactions [with
centroid–centroid distances of 3.561 (2)–3.969 (7) A˚ ] are also observed.
Related literature
For related structures, see: Chantrapromma et al. (2008); Chantrapromma & Fun (2009). For background to non-linear optical materials, see: Dittrichet al.(2003); Nogiet al.(2000); Raimundo et al. (2002); Ruanwas et al. (2010); Sato et al.
(1999).
Experimental
Crystal data
2C16H15N2+C6H4ClO3S
NO3
Mr= 724.21 Triclinic,P1
a= 8.7540 (7) A˚
b= 13.6648 (10) A˚
c= 15.3465 (11) A˚ = 97.206 (1)
= 91.186 (2)
= 99.924 (1)
V= 1792.3 (2) A˚3
Z= 2
MoKradiation = 0.22 mm1
T= 153 K
0.550.470.14 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan
(SADABS; Bruker, 2005)
Tmin= 0.890,Tmax= 0.970
9106 measured reflections 6217 independent reflections 4480 reflections withI> 2(I)
Rint= 0.021
Refinement
R[F2> 2(F2)] = 0.074
wR(F2) = 0.254
S= 1.05 6217 reflections 636 parameters
206 restraints
H-atom parameters constrained
max= 0.87 e A˚
3 min=0.29 e A˚
[image:1.610.315.564.464.635.2]3
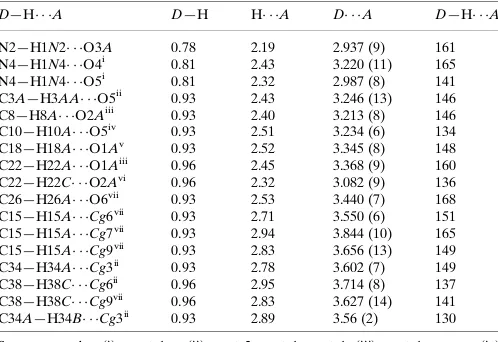
Table 1
Hydrogen-bond geometry (A˚ ,).
Cg3,Cg6,Cg7 andCg9 are the centroids of the C16–C21, C32–C37, N4A/ C30A–C32A/C37Aand C32A–C37Arings, respectively.
D—H A D—H H A D A D—H A
N2—H1N2 O3A 0.78 2.19 2.937 (9) 161 N4—H1N4 O4i
0.81 2.43 3.220 (11) 165 N4—H1N4 O5i 0.81 2.32 2.987 (8) 141 C3A—H3AA O5ii
0.93 2.43 3.246 (13) 146 C8—H8A O2Aiii
0.93 2.40 3.213 (8) 146 C10—H10A O5iv
0.93 2.51 3.234 (6) 134 C18—H18A O1Av
0.93 2.52 3.345 (8) 148 C22—H22A O1Aiii
0.96 2.45 3.368 (9) 160 C22—H22C O2Avi
0.96 2.32 3.082 (9) 136 C26—H26A O6vii 0.93 2.53 3.440 (7) 168 C15—H15A Cg6vii
0.93 2.71 3.550 (6) 151 C15—H15A Cg7vii
0.93 2.94 3.844 (10) 165 C15—H15A Cg9vii 0.93 2.83 3.656 (13) 149 C34—H34A Cg3ii
0.93 2.78 3.602 (7) 149 C38—H38C Cg6ii
0.96 2.95 3.714 (8) 137 C38—H38C Cg9vii
0.96 2.83 3.627 (14) 141 C34A—H34B Cg3ii
0.93 2.89 3.56 (2) 130
Symmetry codes: (i)x;yþ1;z; (ii)xþ2;yþ1;zþ1; (iii)xþ1;y;z; (iv) xþ1;y;zþ1; (v) xþ2;yþ1;z; (vi) x;y1;z; (vii) xþ1;yþ1;zþ1.
Data collection:APEX2(Bruker, 2005); cell refinement:SAINT
(Bruker, 2005); data reduction:SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008); program(s) used to refine structure:SHELXTL; molecular graphics:SHELXTL; software used to prepare material for publication:SHELXTL, PLATON(Spek,
Acta Crystallographica Section E
Structure Reports
Online
The authors thank Prince of Songkla University for generous support and the Universiti Sains Malaysia for the APEX DE2012 grant No. 1002/PFIZIK/910323.
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB7156).
References
Bruker (2005).APEX2,SAINTandSADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
Chantrapromma, S. & Fun, H.-K. (2009).Acta Cryst.E65, o258–o259.
Chantrapromma, S., Kobkeatthawin, T., Chanawanno, K., Karalai, C. & Fun, H.-K. (2008).Acta Cryst.E64, o876–o877.
Dittrich, Ph., Bartlome, R., Montemezzani, G. & Gu¨nter, P. (2003).Appl. Surf. Sci.220, 88–95.
Nogi, K., Anwar, U., Tsuji, K., Duan, X.-M., Okada, S., Oikawa, H., Matsuda, H. & Nakanishi, H. (2000).Nonlinear Optics,24, 35–40.
Raimundo, J.-M., Blanchard, P., Planas, N. G., Mercier, N., Rak, I. L., Hierle, R. & Roncali, J. (2002).J. Org. Chem.67, 205–218.
Ruanwas, P., Kobkeatthawin, T., Chantrapromma, S., Fun, H.-K., Philip, R., Smijesh, N., Padaki, M. & Isloor, A. M. (2010).Synth. Met.160, 819–824. Sato, N., Rikukawa, M., Sanui, K. & Ogata, N. (1999).Synth. Met.101, 132–
133.
supporting information
Acta Cryst. (2013). E69, o1753–o1754 [doi:10.1107/S1600536813030080]
Bis{4-[(E)-2-(1H-indol-3-yl)ethenyl]-1-methylpyridinium}
4-chlorobenzene-sulfonate nitrate
Hoong-Kun Fun, Ching Kheng Quah, Nawong Boonnak and Suchada Chantrapromma
S1. Comment
Organic molecules that exhibit second-order NLO properties usually consist of a framework with delocalized π system, end-capped with either a donor or acceptor substituent or both. Several pyridinium derivatives have been reported to exhibit second-order NLO properties such as single crystals of
1-methyl-4-(2-(4-(dimethylamino)phenyl)-ethynyl)pyridinium p-toluenesulfonate (DAST) and its analogues (Dittrich et al., 2003; Sato et al., 1999). Based on the knowledge that the organic dipolar compounds with extended π systems having terminal donor and acceptor groups are likely to exhibit large hyperpolarizability (β) (Raimundo et al., 2002), we have synthesized several quinolinium derivatives which exhibit NLO properties (Ruanwas et al., 2010). In a similar manner, the title compound (I) was designed and synthesized in order to study for its NLO property. Unfortunately (I) crystallizes in a centrosymmetric P-1 space group which precluded the second-order NLO properties. Herein the crystal structure of (I) is reported.
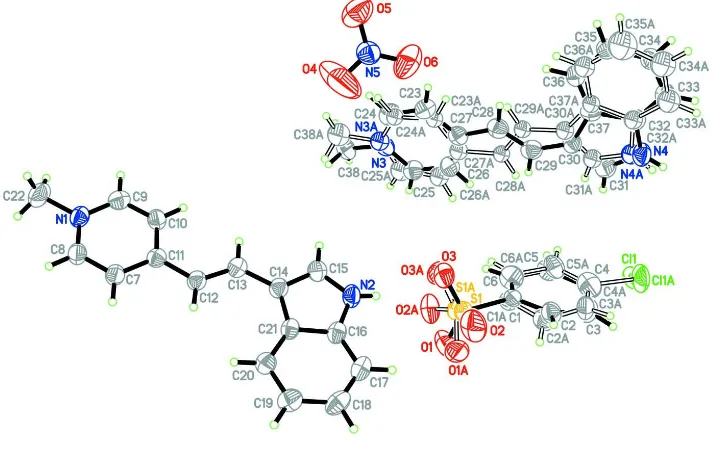
In the crystal structure of (I), the asymmetric unit consists of two C16H15N2+ cations, C6H4ClO3S- and NO3- anions (Fig.
1). One cation [C23–C38/N3/N4] exhibits whole molecule disorder over two sets of sites with a refined site-occupancy ratio of 0.711 (7):0.289 (7). The molecule is disordered in such a way that the ethynyl unit in the major and minor (A) components are related by a 180° rotation. The two cations exist in the E conformation with respect to the ethenyl unit and the torsion angle C11–C12–C13–C14 = -178.8 (3)° for the non-disordered cation, and C27–C28–C29–C30 = 176.8 (6)° and -179.8 (15)° for the major and minor (A) components for the disordered cation. The cations are close to planar with the dihedral angles between the pyridinium and the indole rings being 1.28 (3)° for the non-disordered cation, and 5.62 (3) and 2.45 (3)° for the major and minor components respectively for the disordered cation. The 4-chloro-benzenesulfonate anion also shows whole molecule disorder (Fig. 1) with a 0.503 (6):0.497 (6) site occupancy ratio. Bond lengths of the title compound are comparable to those in related structures (Chantrapromma et al., 2008; Chantrapromma & Fun, 2009)
In the crystal (Fig. 2), the cations are stacked in an antiparallel fashion into columns approximately along the a axis and are further linked to the anions via N—H···O hydrogen bonds and C—H···O interactions (Table 1). C—H···π interactions and π–π interactions were observed with Cg1···Cg2iii = 3.6804 (19) Å, Cg2···Cg3iii = 3.561 (2) Å, Cg2···Cg10vi = 3.969 (7)
Å, Cg2···Cg11vi = 3.949 (7) Å, Cg5···Cg5viii = 3.729 (5) Å, Cg5···Cg8viii = 3.728 (8) Å and Cg8···Cg8viii = 3.741 (11) Å;
Cg1, Cg2, Cg3, Cg5, Cg8, Cg10 and Cg11 are the centroids of N2/C14–C16/C21, N1/C7–C11, C16–C21, N3/C23–C27, N3A/C23A–C27A, C1A–C6A and C1–C6, respectively [symmetry code (viii) = -x, 1 - y, 1 - z].
S2. Experimental
filtered, washed with ether and recrystallized from methanol to give orange single crystals of compound A after several days. The title compound was synthesized by mixing compound A (0.24 g, 0.67 mmol) in hot methanol (30 ml) and silver(I) 4-chlorobenzenesulfonate (0.20 g, 0.67 mmol) in hot methanol (20 ml). The mixture, which turned yellow and cloudy immediately, yielded a gray solid of silver iodide. After stirring the mixture for ca. 30 min, the precipitate of silver iodide was removed and the resulting solution was evaporated to yield an orange solid. Orange blocks of (I) were
recrystalized from methanol solution by slow evaporation of the solvent at room temperature after several days.
S3. Refinement
All H atoms were positioned geometrically and allowed to ride on their parent atoms, with N—H = 0.78, 0.81 and 0.86 Å, CH and Caryl—H = 0.93 Å and Cmethyl—H = 0.96 Å. The Uiso values were constrained to be 1.5Ueq of the carrier atom
for methyl H atoms and 1.2Ueq for the remaining H atoms. A rotating group model was used for the methyl groups. One
[image:4.610.127.485.300.525.2]cation is whole molecule disordered over two sites with refined site occupancies ratio 0.711 (7):0.289 (7), whereas the 4-chlorobenzenesulfonate anion is disordered over two sites with refined site occupancies ratio 0.503 (6):0.497 (6). Similarity and simulation restraints were applied. The displacement ellipsoids of each of the two pairs of atoms i.e. "CL1 C6" and "N3 C38" were restrained to be almost equal.
Figure 1
Figure 2
The crystal packing (involving only the major components of the disordered ions) viewed along the c axis. Hydrogen bonds are drawn as dashed lines.
Bis{4-[(E)-2-(1H-indol-3-yl)ethenyl]-1-methylpyridinium} 4-chlorobenzenesulfonate nitrate
Crystal data
2C16H15N2+·C6H4ClO3S−·NO3−
Mr = 724.21
Triclinic, P1 Hall symbol: -P 1 a = 8.7540 (7) Å b = 13.6648 (10) Å c = 15.3465 (11) Å α = 97.206 (1)° β = 91.186 (2)° γ = 99.924 (1)° V = 1792.3 (2) Å3
Z = 2 F(000) = 756 Dx = 1.342 Mg m−3
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 6217 reflections θ = 2.2–25.0°
µ = 0.22 mm−1
T = 153 K Block, orange
0.55 × 0.47 × 0.14 mm
Data collection Bruker APEXII CCD
diffractometer
Radiation source: sealed tube Graphite monochromator φ and ω scans
Absorption correction: multi-scan (SADABS; Bruker, 2005) Tmin = 0.890, Tmax = 0.970
9106 measured reflections 6217 independent reflections 4480 reflections with I > 2σ(I) Rint = 0.021
θmax = 25.0°, θmin = 2.2°
Refinement Refinement on F2
Least-squares matrix: full R[F2 > 2σ(F2)] = 0.074
wR(F2) = 0.254
S = 1.05 6217 reflections 636 parameters 206 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained w = 1/[σ2(F
o2) + (0.1598P)2 + 0.6516P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max = 0.001
Δρmax = 0.87 e Å−3
Δρmin = −0.29 e Å−3
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq Occ. (<1)
O4 0.7782 (12) 0.1160 (6) 0.5522 (4) 0.262 (4) N1 0.3365 (3) −0.3639 (2) 0.08898 (19) 0.0675 (7) N2 0.7327 (4) 0.2710 (2) 0.1516 (2) 0.0737 (8) H1N2 0.7335 0.3279 0.1670 0.088*
C2A 0.8482 (11) 0.7269 (6) 0.2379 (12) 0.078 (5) 0.503 (6) H2AA 0.9299 0.7149 0.2027 0.094* 0.503 (6) C3A 0.8555 (13) 0.8208 (8) 0.2901 (16) 0.079 (5) 0.503 (6) H3AA 0.9494 0.8651 0.3001 0.095* 0.503 (6) C4A 0.7258 (12) 0.8453 (6) 0.3251 (13) 0.108 (8) 0.503 (6) C5A 0.5943 (14) 0.7713 (7) 0.3256 (16) 0.086 (7) 0.503 (6) H5AA 0.5067 0.7863 0.3539 0.103* 0.503 (6) C6A 0.5948 (9) 0.6747 (5) 0.2832 (12) 0.054 (4) 0.503 (6) H6AA 0.5088 0.6246 0.2851 0.065* 0.503 (6) C10 0.3905 (4) −0.1910 (3) 0.1379 (2) 0.0671 (9)
H10A 0.3681 −0.1366 0.1751 0.080* C9 0.3092 (4) −0.2839 (3) 0.1429 (2) 0.0720 (9) H9A 0.2333 −0.2924 0.1842 0.086* C8 0.4452 (4) −0.3513 (3) 0.0303 (2) 0.0693 (9) H8A 0.4637 −0.4065 −0.0073 0.083* C7 0.5300 (4) −0.2594 (3) 0.0239 (2) 0.0662 (8) H7A 0.6052 −0.2530 −0.0179 0.079* C11 0.5065 (4) −0.1757 (2) 0.0784 (2) 0.0585 (8) C12 0.5998 (4) −0.0784 (2) 0.0709 (2) 0.0616 (8) H12A 0.6757 −0.0759 0.0293 0.074* C13 0.5851 (4) 0.0068 (2) 0.1188 (2) 0.0644 (8) H13A 0.5097 0.0019 0.1606 0.077* C14 0.6697 (4) 0.1055 (2) 0.1148 (2) 0.0612 (8) C15 0.6401 (5) 0.1873 (3) 0.1677 (2) 0.0735 (10) H15A 0.5650 0.1852 0.2097 0.088* C16 0.8290 (4) 0.2470 (2) 0.0854 (2) 0.0625 (8) C17 0.9405 (4) 0.3088 (3) 0.0471 (3) 0.0734 (10) H17A 0.9609 0.3775 0.0649 0.088* C18 1.0202 (4) 0.2658 (3) −0.0180 (3) 0.0835 (12) H18A 1.0965 0.3059 −0.0454 0.100* C19 0.9891 (4) 0.1619 (3) −0.0444 (3) 0.0795 (10) H19A 1.0464 0.1344 −0.0885 0.095* C20 0.8755 (4) 0.0995 (3) −0.0066 (2) 0.0656 (8) H20A 0.8550 0.0309 −0.0249 0.079* C21 0.7926 (4) 0.1428 (2) 0.0601 (2) 0.0568 (7) C22 0.2475 (6) −0.4646 (3) 0.0955 (3) 0.1038 (15) H22A 0.2297 −0.5021 0.0379 0.156* H22B 0.1497 −0.4584 0.1208 0.156* H22C 0.3052 −0.4988 0.1320 0.156*
C35A 0.783 (3) 0.7460 (15) 0.8125 (12) 0.138 (15)* 0.289 (8) H35B 0.8069 0.7156 0.8607 0.166* 0.289 (8) C36A 0.678 (2) 0.6898 (10) 0.7431 (10) 0.071 (7)* 0.289 (8) H36B 0.6433 0.6213 0.7421 0.085* 0.289 (8) C37A 0.628 (3) 0.7422 (8) 0.6764 (13) 0.052 (6)* 0.289 (8) C38A −0.069 (2) 0.2302 (10) 0.3910 (14) 0.103 (8)* 0.289 (8) H38D −0.1751 0.2335 0.4048 0.155* 0.289 (8) H38E −0.0323 0.1828 0.4235 0.155* 0.289 (8) H38F −0.0643 0.2094 0.3291 0.155* 0.289 (8) N5 0.7895 (4) 0.1618 (3) 0.6196 (2) 0.0802 (9)
O5 0.7699 (5) 0.1173 (3) 0.6834 (3) 0.1369 (14) O6 0.8104 (5) 0.2515 (3) 0.6310 (3) 0.1375 (15)
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
C14 0.066 (2) 0.0545 (18) 0.0653 (19) 0.0144 (14) 0.0013 (15) 0.0097 (14) C15 0.090 (3) 0.059 (2) 0.074 (2) 0.0202 (18) 0.0110 (19) 0.0053 (16) C16 0.0616 (19) 0.0528 (17) 0.073 (2) 0.0091 (14) −0.0122 (16) 0.0118 (15) C17 0.067 (2) 0.058 (2) 0.094 (3) 0.0032 (17) −0.0100 (19) 0.0187 (18) C18 0.060 (2) 0.084 (3) 0.107 (3) −0.0039 (19) −0.005 (2) 0.040 (2) C19 0.068 (2) 0.086 (3) 0.091 (3) 0.0222 (19) 0.0137 (19) 0.024 (2) C20 0.062 (2) 0.0618 (19) 0.076 (2) 0.0162 (15) 0.0032 (16) 0.0124 (16) C21 0.0547 (17) 0.0497 (16) 0.0670 (18) 0.0118 (13) −0.0075 (14) 0.0104 (13) C22 0.117 (4) 0.071 (3) 0.111 (3) −0.025 (2) −0.003 (3) 0.021 (2) N3 0.064 (4) 0.093 (6) 0.073 (5) 0.022 (3) 0.006 (2) −0.007 (3) N4 0.105 (5) 0.055 (3) 0.092 (4) 0.012 (3) −0.016 (3) 0.008 (3) C23 0.081 (4) 0.081 (4) 0.082 (4) 0.013 (4) 0.005 (3) 0.022 (4) C24 0.070 (6) 0.088 (6) 0.100 (6) 0.014 (3) 0.014 (4) 0.016 (4) C25 0.078 (5) 0.074 (4) 0.081 (4) 0.025 (4) 0.004 (3) 0.016 (4) C26 0.084 (5) 0.073 (4) 0.077 (5) 0.019 (3) 0.009 (4) 0.004 (3) C27 0.062 (3) 0.073 (4) 0.067 (4) 0.019 (3) 0.011 (3) 0.003 (3) C28 0.084 (4) 0.084 (4) 0.071 (3) 0.027 (3) 0.005 (3) 0.009 (3) C29 0.086 (4) 0.081 (4) 0.075 (3) 0.024 (3) 0.013 (3) 0.016 (3) C30 0.070 (3) 0.058 (3) 0.070 (3) 0.018 (3) 0.001 (3) 0.008 (3) C31 0.097 (5) 0.072 (4) 0.083 (4) 0.019 (4) −0.006 (3) 0.017 (4) C32 0.067 (8) 0.070 (8) 0.080 (10) 0.018 (3) 0.011 (3) 0.011 (3) C33 0.063 (4) 0.066 (5) 0.081 (5) 0.006 (3) 0.002 (4) 0.008 (3) C34 0.066 (4) 0.068 (4) 0.072 (4) 0.006 (3) −0.008 (2) 0.011 (3) C35 0.079 (4) 0.064 (3) 0.065 (3) 0.019 (3) −0.005 (3) 0.011 (3) C36 0.064 (4) 0.064 (4) 0.071 (4) 0.010 (2) 0.003 (3) 0.000 (3) C37 0.062 (4) 0.073 (4) 0.057 (4) 0.023 (3) 0.013 (3) 0.010 (3) C38 0.073 (4) 0.094 (4) 0.082 (4) 0.012 (3) −0.008 (3) −0.015 (4) N5 0.084 (2) 0.084 (2) 0.075 (2) 0.0217 (18) −0.0086 (17) 0.0104 (18) O5 0.143 (3) 0.116 (3) 0.163 (4) 0.028 (2) 0.061 (3) 0.048 (3) O6 0.129 (3) 0.094 (3) 0.195 (4) 0.014 (2) −0.024 (3) 0.049 (3)
Geometric parameters (Å, º)
O4—N5 1.133 (6) N4—C32 1.372 (6)
N1—C8 1.328 (5) N4—H1N4 0.8081
N1—C9 1.345 (5) C23—C24 1.366 (8) N1—C22 1.476 (5) C23—C27 1.429 (9) N2—C15 1.336 (5) C23—H23A 0.9300 N2—C16 1.377 (5) C24—H24A 0.9300 N2—H1N2 0.7838 C25—C26 1.376 (11) Cl1—C4 1.842 (5) C25—H25A 0.9300 S1—O3 1.342 (4) C26—C27 1.409 (10)
S1—O1 1.425 (5) C26—H26A 0.9300
S1—O2 1.427 (7) C27—C28 1.468 (7) S1—C1 1.773 (4) C28—C29 1.318 (6)
C1—C6 1.352 (8) C28—H28A 0.9300
C2—H2A 0.9300 C30—C31 1.391 (9) C3—C4 1.343 (9) C30—C37 1.445 (8)
C3—H3A 0.9300 C31—H31A 0.9300
C4—C5 1.395 (8) C32—C33 1.377 (6) C5—C6 1.397 (8) C32—C37 1.412 (7)
C5—H5A 0.9300 C33—C34 1.362 (8)
C6—H6A 0.9300 C33—H33A 0.9300
Cl1A—C4A 1.842 (5) C34—C35 1.363 (10) S1A—O3A 1.342 (4) C34—H34A 0.9300 S1A—O1A 1.425 (5) C35—C36 1.443 (9) S1A—O2A 1.427 (7) C35—H35A 0.9300 S1A—C1A 1.773 (4) C36—C37 1.421 (8) C1A—C6A 1.351 (8) C36—H36A 0.9300 C1A—C2A 1.365 (7) C38—H38A 0.9600 C2A—C3A 1.415 (10) C38—H38B 0.9600
C2A—H2AA 0.9300 C38—H38C 0.9600
C3A—C4A 1.343 (10) N3A—C25A 1.324 (9) C3A—H3AA 0.9300 N3A—C24A 1.352 (7) C4A—C5A 1.395 (8) N3A—C38A 1.468 (6) C5A—C6A 1.397 (8) N4A—C31A 1.326 (8) C5A—H5AA 0.9300 N4A—C32A 1.372 (6)
C6A—H6AA 0.9300 N4A—H1N4 1.1137
C10—C9 1.356 (5) N4A—H2N4 0.8600 C10—C11 1.386 (5) C23A—C24A 1.367 (8) C10—H10A 0.9300 C23A—C27A 1.429 (9)
C9—H9A 0.9300 C23A—H23B 0.9300
C22—H22A 0.9600 C36A—H36B 0.9300 C22—H22B 0.9600 C38A—H38D 0.9600 C22—H22C 0.9600 C38A—H38E 0.9600 N3—C25 1.324 (9) C38A—H38F 0.9600 N3—C24 1.352 (7) N5—O6 1.197 (5) N3—C38 1.468 (6) N5—O5 1.216 (5) N4—C31 1.326 (8)
C17—C18—H18A 119.5 C33A—C34A—C35A 121.8 (5) C19—C18—H18A 119.5 C33A—C34A—H34B 119.1 C20—C19—C18 121.7 (4) C35A—C34A—H34B 119.1 C20—C19—H19A 119.2 C34A—C35A—C36A 120.4 (4) C18—C19—H19A 119.2 C34A—C35A—H35B 119.8 C19—C20—C21 117.9 (3) C36A—C35A—H35B 119.8 C19—C20—H20A 121.1 C37A—C36A—C35A 118.0 (5) C21—C20—H20A 121.1 C37A—C36A—H36B 121.0 C20—C21—C16 118.6 (3) C35A—C36A—H36B 121.0 C20—C21—C14 135.0 (3) C32A—C37A—C36A 117.3 (6) C16—C21—C14 106.4 (3) C32A—C37A—C30A 107.4 (6) N1—C22—H22A 109.5 C36A—C37A—C30A 135.4 (5) N1—C22—H22B 109.5 N3A—C38A—H38D 109.5 H22A—C22—H22B 109.5 N3A—C38A—H38E 109.5 N1—C22—H22C 109.5 H38D—C38A—H38E 109.5 H22A—C22—H22C 109.5 N3A—C38A—H38F 109.5 H22B—C22—H22C 109.5 H38D—C38A—H38F 109.5 C25—N3—C24 120.4 (6) H38E—C38A—H38F 109.5 C25—N3—C38 123.1 (7) O4—N5—O6 123.5 (6) C24—N3—C38 116.5 (7) O4—N5—O5 118.0 (6) C31—N4—C32 109.5 (6) O6—N5—O5 118.3 (4)
Hydrogen-bond geometry (Å, º)
Cg3, Cg6, Cg7 and Cg9 are the centroids of the C16–C21, C32–C37, N4A/C30A–C32A/C37A and C32A–C37A rings, respectively.
D—H···A D—H H···A D···A D—H···A
N2—H1N2···O3A 0.78 2.19 2.937 (9) 161 N4—H1N4···O4i 0.81 2.43 3.220 (11) 165
N4—H1N4···O5i 0.81 2.32 2.987 (8) 141
C3A—H3AA···O5ii 0.93 2.43 3.246 (13) 146
C8—H8A···O2Aiii 0.93 2.40 3.213 (8) 146
C10—H10A···O5iv 0.93 2.51 3.234 (6) 134
C18—H18A···O1Av 0.93 2.52 3.345 (8) 148
C22—H22A···O1Aiii 0.96 2.45 3.368 (9) 160
C22—H22C···O2Avi 0.96 2.32 3.082 (9) 136
C26—H26A···O6vii 0.93 2.53 3.440 (7) 168
C15—H15A···Cg6vii 0.93 2.71 3.550 (6) 151
C15—H15A···Cg7vii 0.93 2.94 3.844 (10) 165
C15—H15A···Cg9vii 0.93 2.83 3.656 (13) 149
C34—H34A···Cg3ii 0.93 2.78 3.602 (7) 149
C38—H38C···Cg6ii 0.96 2.95 3.714 (8) 137
C38—H38C···Cg9vii 0.96 2.83 3.627 (14) 141
C34A—H34B···Cg3ii 0.93 2.89 3.56 (2) 130