ANTIFUNGAL SCREENINGOF NITROGEN AND SULPHUR DONOR
ATOM CONTAINING LIGAND
3,3’-THIODIPROPIONICACID-BIS(5-AMINO-1,3,4-THIODIAZOLE-2-THIOL) AND ITS Mn(II),Co(II)
COMPLEXES: SYNTHESIS, SPECTRSCOPIC APPROCH,
MOLECULAR MODELING
Sulekh Chandra*1, Seema Gautam1, Amit Kumar2
1
Department of Chemistry, Zakir Husain Delhi College (University of Delhi), JLN Marg,
New Delhi-110002, India
2
Department of Chemistry, DayalSingh College (University of Delhi), New
Delhi-110003, India
ABSTRACT
A novel series ofMn(II)and Co(II)complexes were synthesized by
condensation reaction of nitrogen and sulphur donor atom containing
ligand 3,3’-thiodipropionicacid-bis (5-amino-1,3,4-thiodiazole-2-thiol)
(L) and corresponding metal salt in 1:1 ratio. This multi donor ligand
was synthesized by conventional condensation reaction between
3,3’-thiodipropionic acid and 5-amino-1,3,4-thiodiazole-2-thiol in 1:2 ratio.
The mode of bonding of ligand toto metal ion and geometry of the
synthesized complexes were inferred through elemental analysis, 1 H-NMR, Mass, IR, molar conductance, magnetic susceptibility,
electronic spectra, epr, and molecular modeling. On the basis of above
studies, octahedral geometry was assigned for Mn(II) andCo(II)
complexes. The in vitroantifungal activityofligand and its Mn(II)and Co(II) complexes was studied. Fungi C.krusei, C.albicans, C.tropicalisand C. parapsilos were takenand well diffusion method was applied to study antifungal activity. A comparative study ofinhibition
values indicated that complexes exhibited higher activity than the ligand.
KEY WORDS: 3,3’-thiodipropionic acid; 1H-NMR;electronic spectra; epr; molecular modeling; C. krusei; C. albicans; C. tropicalis; C. parapsilos; well diffusion method.
Volume 3, Issue 7, 576-595. Research Article ISSN 2277 – 7105
Article Received on 06 July 2014,
Revised on 30 July 2014, Accepted on 24 August 2014
*Correspondence for
Author
Dr. Sulekh Chandra
INTRODUCTION
The quick development of infectious disease resistance to most ofthe known drugs is
becoming a serious health disaster [1]. One probable long-term key is the improvement of used drugs that act on targeted microorganismi.e. Bacteria, fungi and virus [2,3]. The investigation for novel antimicrobial drugis one of the mostdemanding tasks to the chemist [4]. Transition metal complexes have received a considerable attention in last many years, due to their
interesting characteristics in biological field [5]. Ligands containing nitrogen, oxygen, and sulphur donor atoms have wide variety to coordinateto metal ions,were published in
biological field[6].The preparation of a new ligand, which contain donor atoms is most importantstep in the development of metal complexes which exhibitunique properties and
novel reactivity[7,8].1,3,4-Thiadiazoles attract great attention after the discovery of sulfa drugs
and their potent representative bearing this heterocyclic ring[9]. Several1,3,4-thiadiazoles moiety containing ligands were found biologically active, e.g., they showed antimicrobial activity[10,11],anticancer[12], antiviral[13],anti-inflammatory, antioxidant[14]and antitumor[15] etc. In present work, we report the synthesis procedure and structural characterization of nitrogen
and sulphur donor atoms containing ligand
3,3’-thiodipropionicacid-bis(5-amino-1,3,4-thiodiazole-2-thiol)(L), and its Mn(II) and Co(II) complexes. In addition, we have
evaluatedin vitro antifungal activity of synthesized ligand and its complexesagainst some fungi strain.
MATERIAL AND METHODS Materials
All reagents were used without further purification and used as received. 3,3’-thiodipropionic
acid and 5-amino-1,3,4-thiodiazole-2-thiol, were of AR grade and procured from Alfa Aesar,
Heysham, England and Sigma Aldrich, Bangalore, India. All used solvents were of
spectroscopic grade and obtained from Merck or Aldrich. The metal salts of MnCI2.4H2O,
CoCI2.6H2O, Mn(CH3COO)2, and Co(CH3COO)2, were purchased from Merck.
Analytical And Physical Methods
Elemental analyses (CHN) were carried out on a Carlo-Erba 1106 elemental analyzer. 1 H-NMR spectra were recorded using a Bruker Advanced DPX-300 spectrometer using
DMSO-d6 as a solvent and TMS as an internal solvent, at IIT Delhi. Molar conductance was
measured on the ELICO (CM82T) conductivity bridge. Magnetic susceptibility was
were recorded using a FT-IR spectrum BX-II spectrophotometer on KBr disks in the wave
number ranged400-4000 cm-1. Electronic spectral studies were done on a Shimadzu model UV-1200 spectrophotometer. ESR spectra of complexes were recorded as polycrystalline
sample in DMSO solution at liquid nitrogen temperature (LNT) on E4-ESR spectrometer
using the DPPH as the g-maker.
Molecular Modeling
Molecular modeling of the proposed structure of ligand and its complexes wasmodelledby
using Gauss View 5.0 at Semi empirical PM6 level.In order to obtain the structural
information for the ligand and its complexes, Geometry optimisation was carried out for both,
the ligand and complexes, [M(L)Cl]Cl and [M(L)CH3COO]CH3COO; [M = Mn(II) and
Co(II)] by implementation of semi empirical (PM6) method of the Gaussian 09 W package, in the gas phase.
Antifungal Screening
The in vitro evaluation of antifungalactivity was carried out by using well diffusion method[16]. The purpose of thescreening program was to provide antimicrobial efficienciesof the investigated compounds. The prepared compoundswere tested against some fungi
Candida albicans, Candida parapsilosis, Candidiakrusei, and Candida tropicalis which were obtained from Microbiology Department, Medical College, Subharti Vivekanand University,
and Meerut. Allthe fungus strain was maintained in Saboraud Dextrose broth (SDB) at
-150C.
Well Diffusion Method
In well diffusion method, 1.0 mL of antibacterial drug (Tetracyclin) was added to50 mL of
Saboraud Dextrose Agar media. The purpose to mix tetracycline was to kill the bacterial stain
if present in SDA. In laminar flow, prepared media was poured into 120 mm petri plates and
allowed to cool at room temperature. Wells (6 mm in diameter) were cut in plates using
sterile tubes. Sterilized swabs were dipped into the nutrient broth of fungi, used to rub the
surface of the plates. Then, tested compound at desired concentration (1000, 750, 500, 250
ppm) were poured into prepared well. The plates were sealed with Parafilm, and incubated at
temperature 22±290C for 48 hrs. DMSO was used as control under the same conditions for each micro organism. Antifungal drug Nystatin was used as a standard drug for comparison
purpose. The growth of fungi (mm) in each plate was measured diametrically and inhibition
C = Radial diameters of the colony in control, T = Radial diameter of the colony in test
compound.
Experimental Details
Synthesis of Ligand 3, 3’-Thiodipropionicacid-Bis(5-Amino-1,3,4-Thiodiazole-2-Thiol)(L)
5-amino-1,3,4-thiodiazole-2-thiol (2 mol) was dissolved in 15 mL EtOH, and this ethanolic
solution was added dropwise into an ethanolic solution of 3,3’ thiodipropionic acid (1 mol),
and this reaction solution was refluxed at 80-850C for 5 hrs(Fig 1).On cooling, cream-colored precipitate was formed. This precipitate was filtered off, washed with cold ethanol, and
distilled water. After proper washing, resulting product was dried under vacuum over P4O10.
S O
NH
S N
N
SH
O
HN
S
N
N
HS S
O
OH
O
OH H2N S
N N
SH
[image:4.595.128.471.311.518.2]3,3'thiodipropionic acid 5-amino-1,3,4-thiodiazole-2-thiol
Fig. 1 Synthesis of Ligand Synthesis of complexes
Synthesized ligand (0.001 mol, 0.408g) was dissolved in 20 mL EtOH in a 100 mL
round-bottom flask. Hot ethanolic solution of the metal salt (0.001 mol) [MnCI2.4H2O,
CoCI2.6H2O,Mn(CH3COO)2, and Co(CH3COO)2] were mixed with hot ethanolic solution of
ligand and content was further refluxed for 10-12 hrs at temp 85-900C. pH (5-7) was adjusted by adding of 2-3 drops of aqueous ammonia. The corresponding coloured complexes were
separated out by filtration, washed thoroughly with ethanol, distilled water and dried under
Table 1Analytical and physical data of ligand and its transition metal complexes
Compound Mol. Wt. Melting Point Colour
Molar conduct-
ance
Magnetic moment
Elemental Analysis
C H N M
Ligand (L) C10H12N6O2S5 408 180 White - - 29.41 (29.40) 2.94 (2.92) 20.58 (20.58) -
[Mn(L)Cl]Cl 533.9 >250 Light Brown 158 5.92 22.47 (22.45) 2.24 (2.25) 15.73(15.70) 10.28 (10.25)
[Mn(L)OAc]OAc 580.9 Light Creemish 136 5.95 28.92 (28.91) 3.09 (3.05) 14.46 (14.48) 9.45 (9.43)
[Co(L)Cl]Cl 569.9 >250 Pink 120 4.79 22.30 (22.24) 2.23 (2.23) 15.61 (15.60) 10.94 (10.99)
[Co(L)OAc]OAc 584.9 Rust Red 206 4.92 28.72 (28.70) 3.07 (3.03) 14.36 (14.34) 10.07 (10.07)
[image:5.842.43.821.295.455.2]CH3COO is written as OAc
Table 2 Characteristic IR Bands (Cm-1) of Ligand And Its Complexes
Compound ν(NH) ν (C-S-C) ν(C=O) ν(M-N) ν(M-Cl) Bands due to anion
Ligand (L) C10H12N6O2S5 1552 754 1609 - -
[Mn(L)Cl]Cl 1507 733 1609 445 297 Band observed at307-270-cm
-1
indicated the presence of M-Cl bond
[Mn(L)OAc]OAc 1507 689 1606 445
νas(OAc) = 1401, νs (OAc) = 1290, Δν= 111
cm-1 indicated monodentate nature of acetategroup
[Co(L)Cl]Cl 1508 703 1609 433 305 Band observed at 307-270cm
-1
indicated the presence of M-Cl bond
[Co(L)OAc]OAc 1515 744 1609 442 νas(OAc) = 1410, νs(OAc) =1302,Δν = 108 cm
-1
RESULT AND DISCUSSION
The analytical data of complexes is given in Table 1, which indicated that all complexes had
the general composition [M(L)X2 where M = Mn(II), Co(II), L =
3,3’-thiodipropionic-bis(5-amino-1,3,4-thiodiazole-2-thiol), and X = Cl-, CH3COO-. Allthe complexes were sparingly
soluble in common organic solventssuch as ethanol and methanol but highly soluble in
DMSO and DMF. The molar conductance values ofcomplexes in 10−3 molar DMSO solution was in the range120-206 Ω−1cm2 mol−1, which revealed the 1:1 electrolytic nature of metal complexes. On the basis of molar conductance complexes are formulated as [M(L)X]X.
S O NH S N N SH O HN S N N HS HS O HN S N N HS 220 O HN S N N HS 188 S N N HS 117 408 -188 -C5N3S2H6
-S -32 S N N -SH -33
-71 -C3H5NO
-1 H S O NH S N N S O HN S N N HS 407 -32 -S S O NH S N N O HN S N N HS 375
-84 -C2N2S
S NH O HN S N N HS 291 -CHNO S O HN S N N HS 248 -28
-C2H4 S O HN S N N HS 220 -32 -S O HN S N N HS 220 -28 -C2H4
[image:6.595.84.513.245.744.2]O HN S N N HS 160 -CHNO -43 S N N HS S N N 84 117 -43 -SH -33
Mass spectra
The mass spectrum showed a molecular ion peak at m/z 409 which was equivalent to its
molecular weight [L+H]+. The peaks at m/z = 375 and 220 are due to the -SH group and the hetero cyclic ring (C5N3S2H6). [17]. Fig. 2 represented the main fragmentation paths of ligand.
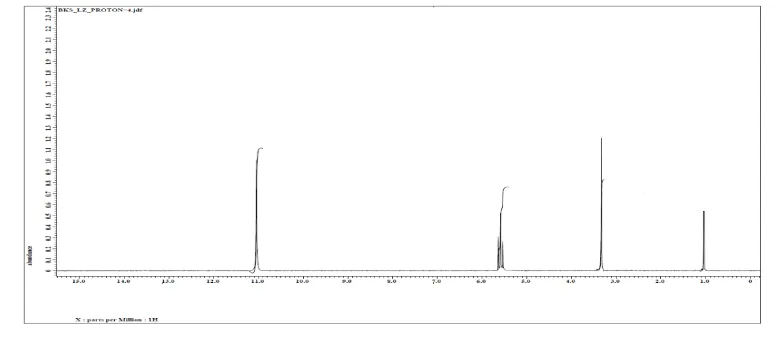
H-NMR spectra
H-NMR spectrum of ligand [3,3-thiodipropionicacid-bis(5-amino-1,3,4-thiodiazole-2-thiol)]
(Fig. 3) in DMSO-d6 showed following signals: two multiple’s at δ 5.7-5.9 ppm (m, 12H,
6CH2) due to the protons of six methylene groups, broad signal atδ 9.6 ppm (s, 2H, 2NH),
corresponding to the two protons of two NH groups [18]. Signal at δ 3.0-3.5 for (s, 2H, 2SH)
[19]
[image:7.595.100.487.298.470.2].
Fig. 3.1H-NMR Spectrum of The Ligand IR spectra
Selected IR bands of the ligand and its complexes are listed in Table 2. The IR spectrum of
the pantadentate ligand displayed bands at 1609, and 1552 cm-1, which might be assigned to the ν(C=O) of amide group, and stretching vibration of ν(NH) group[20]. The band appearing at 754 cm-1 in the spectrum corresponded to the ν(C-S) stretching vibration. The bands which appeared at 1552 cm-1, and 768 cm-1 in IR spectrum of ligand, shifted downward in all metal complexes which indicated that the nitrogen atoms of NH groups and sulphur atoms of C-S-C
group coordinated to the central metal atom. The coordination behaviour of ligand through
nitrogen, was also confirmed by the appearance of new IR bands in the spectra of metal
complexes in 459-420 cm-1 range. These bands might be assignedto ν(M-N) stretching vibrations, respectively[21]. However, the band corresponding to the C=O group remains almost unchanged on complexation, which indicateed that oxygen atom of carbonyl group
metal ion in pantadentate fashion (N2S3)[22].The IR spectra of metal complexes also displayed
the bands due to anions. The chloro complexes showed band in the region 307-270 cm-1, which corresponded to the presence of ν(M-Cl) bond[23]. The acetato complexes showed IR bands in the region 1438-1401 cm-1 and 1330-1290cm-1 due to νas(OAc) and νs(OAc)
stretching vibrations, respectively. The value of Δνlies in the range of 204-187 cm-1
, which
suggested the unidentatebehaviour of acetate ion[24,25].
Magnetic Moment
The magnetic moment observed for Mn(II) and Co(II) complexes lies in the range 5.92-5.95
BM, and 4.79-4.92 BM corresponding to presence of five and three unpaired electrons,
respectively[26,27]. The observed magnetic moments of Mn(II) and Co(II) complexes are given in Table 1.
Electronic Spectra
The electronic spectra of Mn(II) complexes displayed the absorption bands in the range of
17,850-18,518, 23,034-24,096, 26,809-28,188 and 34,482-37,735 cm-1. These transitions may be assigned to the 6A1g→4T1g (4G), 6A1g→4Eg, 4A1g (4G), 6A1g→4Eg (4D) and 6A1g→4T1g (P) [28]
. These transitions revealed the octahedral geometry for the Mn(II) complexes. The
electronic spectra of Co(II) complexes under study displayed the absorption bands in the
range of 9310-9940, 16,290-19,143 and 20,510-22,339cm-1. These transitions may be assigned to the 4T1g(F)→4T2g (F), 4T1g (F)→4A2g (F), and 4T1g (F)→4T1g (P). The transitions
were corresponding to the octahedral geometry of the Co(II) complexes[29].
Ligand Field Parameter
The ligand field parameters like B, C (Racah parameters), 10 Dq, covalency factor and LFSE were calculated for complexes. Slater Condon-shortly repulsion parameters F2 and F4 are
relatedto Racah parameters B and C as: B = F2-5F4 and C=35F4, The electron-electron
repulsion in the complexes was more than in the free ion, resulting in an increased distance
between electrons, and thus, affects the size of the orbital. Onincreasing delocalization, the
parameter βand hxsuggested that thecomplexes, reported here, had appreciable ionic
[image:9.595.102.510.146.673.2]character[30,31].The calculated values of the ligand field parametersare given in Table 3.
Table 3 Electronic spectral data of the complexes and ligand field parameters
Compound λmax(cm-1) Dq B’ ᵝ C F2 F4
LFSE (KJ mol-1)
hx
[Mn(L)Cl]Cl 17852, 23035, 26810,
34485 1785 401 0.51 3805 941 108 - 7
[Mn(L)OAc]OAc 18510, 24090, 28180,
37730 1851 521 0.66 3776 1056 107 - 4.8
[Co(L)Cl]Cl 9560, 18621, 23371,
37735 956 887 0.79 - - - 137 -
[Co(L)OAc]OAc 9945, 17600, 22271,
37593 994 746 0.66 - - - 142 -
Fig.4. UV-Visible Spectra of (A) [Mn(L)Cl]Cl(B) [Co(L)Oac]Oac Electron Spin Resonance (ESR)
completely removed by a magnetic field. The broad signalin the polycrystalline ESR spectra
of Mn(II) complexes is attributed to forbidden transitions, where M=±1 (M= electronspin quantum number). The broadening of spectra is due to immobilization of Mn(II) ion in the
ligand results because the rotational motion of Mn(II) is highly restricted. Anotherorigin of
line broadening is due to spin relaxation, which is temperature dependent. All the Mn(II)
complexes, understudy, shows isotropic ESR spectra (Fig.6), when recorded aspolycrystalline
sample. The X-band ESR spectra of the Co(II) complexes were recorded at liquid nitrogen
temperature in polycrystalline form. The ESR spectra of Co(II) complexes with giso =
2.01-2.04 correspond to the tetragonal symmetry around the Co(II) ion (Table 4) [32,33]. As a
consequence of the fast spin-relaxation time of high-spin Co(II)ion, the signals are observed
only at low temperature.
Table 4 ESR Spectral Data of The Mn(II) And Co(II) Complexes
Complexes g‖ g⊥ giso
[Mn(L)Cl]Cl - - 2.19
[Mn(L)OAc]OAc
- - 2.18
[Co(L)Cl]Cl 2.32 1.89 2.01
[Co(L)OAc]OAc 2.40 1.86 2.04
(b)
(c)
Fig.5 ESR Spectra of (A) [Mn(L)Cl]Cl, (B) [Mn(L)Oac]Oac, (C) [Co(L)Oac]Oac Molecular Modelling
With an objective to obtain the structural information for the ligand and its complexes,
geometry optimisation was carried out for all i.e. ligand, [M(L)Cl]Cl and [M(L)OAc]OAc,
where M = Mn(II) and Co(II), at semiempirical (PM6) level of theory using the Gaussian 09W package, in gas phase. All complexes were found to possess six coordinated octahedral geometry with slightly distortion. In these complexes, one axial position was engaged by
sulphur atom (S1) of ligandFig. 6, whereas another axial position was being engaged by Cl
-ion in [M(L)Cl]Cl complexes and by O-atom of the OAc -ion in [M(L)OAc]OAccomplexes.
The two equatorial positions were occupied by donor N-atoms (N16, N18) and the other two
positions by sulphur atoms (S28, S29) of ligand. The axial Mn-S bond distance for
[Mn(L)Cl]Cl and [Mn(L)OAc]OAc complexes originated at2.361, and 2.236 Å, respectively.
Axial Mn-X distance(X=Cl-/OAc) for [Mn(L)Cl]Cl and [Mn(L)OAc]OAc complexes originate at 2.037, 1.866 Å respectively. Bond distances in four equatorial positions
comprising, Mn-N16, Mn-N18, Mn-S28, and Mn-S29 bonds, were 2.150, 2.070, 2.505, and
case of [Mn(L)OAc]OAccomplex, was found to be 2.026, 2.020, 2.343, and 2.319Å,
respectively.The axial Co-S bond distance for [Co(L)Cl]Cl and [Co(L)OAc]OAccomplexes
was found to be 2.10 and 1.95Å, respectively. Axial Co-X (X = Cl-/OAc) distance for [Co(L)Cl]Cl and [Co(L)OAc]OAccomplexes was found to be 2.371, 1.895 Å respectively.
Bond distances in four equatorial positions comprising, Co-N16, Co-N18, Co-S28, and Co-S29
bonds is found as 1.991, 2.087, 2.087, and 2.155Å, respectively in case of [Co(L)Cl]Cl,
whereas corresponding values in [Co(L)OAc]OAcare 2.11, 2.11, 2.11, and 2.10Å,
respectively Table 5.In the case of [Co(L)OAc]OAc the S27-Mn-N16, S33-Mn-N18, S27-Mn-S33
and N16-Mn-N18 bond angles, in the equatorial plane of the coordination sphere, were
80.49º,80.39º,116.450 and 81.50, respectively. Similar trends were also observed for other complexes. The S1-M-X (X = Cl/OAc) bond angle in [Mn(L)Cl]Cl, [Mn(L)OAc]OAc,
[Co(L)Cl]Cl and [Co(L)OAc]OAcwas bent at 149.050, 155.870, 134.420 and 1650Table 6. These deviations in bond angles in the coordination sphere, resulted in distortion from a
[image:12.595.70.530.403.765.2]regular octahedral geometry in Mn(II) and Co(II) complexes.
Table 5 Optimized Geometry of Legend and Its Complexes (Bond Length In Angstrom) Paremeters Ligand [Mn(L)Cl]Cl [Co(L)Cl]Cl [Co(L)OAc]OAc [Co(L)OAc]OAc
S1-C8 1.825 1.811 1.878 1.861 1.879
C8-C11 1.516 1.532 1.524 1.524 1.522
C11-C15 1.522 1.507 1.509 1.497 1.506
C15-O21 1.216 1.199 1.206 1.198 1.208
C15-N18 1.398 1.498 1.493 1.522 1.477
N18-C25 1.742 1.432 1.432 1.440 1.422
S29-C25 1.805 1.828 1.783 1.797 1.783
C25-N32 1.309 1.358 1.347 1.323 1.357
N32-N33 1.359 1.325 1.348 1.362 1.342
N33-C26 1.322 1.361 1.336 1.324 1.334
C26-S36 1.711 1.705 1.785 1.780 1.787
S36-H37 1.355 1.356 1.350 1.349 1.350
S1-C2 1.825 1.811 2.476 1.858 1.881
C2-C5 1.516 1.520 1.522 1.523 1.521
C5-C14 1.522 1.507 1.506 1.496 1.505
C14-C16 1.398 1.498 1.485 1.520 1.471
C14-O20 1.216 1.199 1.208 1.199 1.209
N16-C23 1.742 1.814 1.431 1.438 1.422
S28-C23 1.805 1.814 1.811 1.798 1.786
S28-C22 1.799 1.780 1.777 1.787 1.768
C22-S34 1.711 1.760 1.773 1.773 1.773
S34-H35 1.355 1.332 1.354 1.357 1.353
C22-N31 1.322 1.339 1.349 1.327 1.338
N30-C23 1.309 1.342 1.375 1.328 1.360
S1-M38 - 2.361 2.103 2.236 2.122
M38-X39 - 2.037 2.371 1.866 1.855
M38-N18 - 2.070 2.087 2.029 2.146
M38-N16 - 2.150 1.991 2.026 2.184
M38-S28 - 2.505 2.087 2.343 2.127
M38-S29 - 3.642 2.155 2.319 2.131
Table 6 Optimized Geometry of Ligand and Its Complexes (Bond Angle In Degree) Parameters Ligand [Mn(L)Cl]Cl [Co(L)Cl]Cl [Co(L)OAc]OAc [Co(L)OAc]OAc
C2-S1-C8 103.58 103.93 97.72 97.77 94.80
C8-C11-C15 113.67 113.41 109.43 111.37 110.38
O21-C15-N18 119.10 115.25 115.83 113.32 115.48
N18-C25-S29 104.77 113.58 112.98 107.72 116.70
C25-N32-N33 115.33 112.36 112.33 111.62 112.39
N33-C26-S29 111.54 122.22 111.69 110.59 111.60
C26-S36-H37 99.14 101.05 96.61 97.40 96.46
S1-C8-C11 107.47 107.31 105.11 108.01 108.02
S1-C2-C5 107.47 107.99 107.55 109.30 109.50
C2-C5-C14 113.67 113.79 109.38 112.18 110.69
C14-N16-S28 118.81 118.68 117.75 119.67 117.42
S28-C23-N30 111.52 112.29 112.05 114.14 112.73
C23-N30-N31 115.33 113.09 112.54 111.49 112.25
N30-N31-C22 114.77 116.76 116.07 116.21 115.65
S28-C22-S34 123.59 123.31 122.40 121.38 121.44
C22-S34-H35 99.14 97.33 96.06 96.50 96.35
S1-M38-X39 - 148.81 134.42 120.99 118.56
N16-M38
-N18
- 127.62 122.87 147.96 128.58
S29-M38-S28 - 77.48 73.39 64.92 72.53
S29-M38-X39 - 107.60 104.47 125.26 125.01
S28-M38-S39 - 115.22 119.49 133.26 129.91
(b) (d)
(d) (e)
Fig. 6 Optimized structure of ligand and complexes (a) ligand (with symbol and numbering) (b) [Mn(L)Cl]Cl, (c) [Mn(L)OAc]OAc, (d) [Co(L)Cl]Cl, (e) [Co(L)OAc]OAc
Fungicidal screening
The antimicrobial screening data showed that ligand and its transition metal complexes
exhibitedantifungalactivity and it is important to note that themetal chelates exhibited more
inhibitory effects than the parent ligand(Fig. 5). The increased activity of complexes can be
explainedon the basis of chelation theory[34].On chelation, the polarity of the metal ion will be
reduced to a greater extent dueto the overlap of the ligand orbital and partial sharing of
positive charge of metal ion withdonor groups[35].Further, it increases the delocalization of the π electrons over thewhole chelate ring and enhances the lipophilicity of the complex. This
increasedlipophilicity enhances the penetration of the complexes into lipid membrane and
thusblocks the metal binding sites on enzymes of microorganisms. These metalcomplexes
[image:14.595.74.514.77.450.2]restricts further growth of the organism. The variation in the activityof different complexes
against different organisms depend either on the impermeability ofthecells of the microbes or
difference in ribosomes of microbial cells.It has also beenproposed that concentration plays a
vital role in increasing the degree of inhibition; as the concentration of tested compounds was
increased the activityalso increased. The activity data of tested compounds is given in Table
[image:15.595.60.534.242.616.2]7.
Table 7 Antifungal screening result of ligand and its complexes
NA means no activity
Compound Concentration (ppm)
Candida albicans
Candida
parapsilosis Candidiakrusei
(a)
(b)
[image:16.595.146.446.79.688.2](c)
S
NH
S N
N
SH
HN
S
N
N
HS M
X
O O
X
Fig. 8. Proposed Structure of complexes where M = Mn(II), Co(II), and X = Cl-, CH3COO
-CONCLUSION
Continuing the synthetic strategies,ligand (N2S3 type)
3,3’-thiodipropionicacid-bis(5-amino-1,3,4-thiodiazole-2-thiol)and its Mn(II) and Co(II) complexeswere synthesized.The ligand
interactedtoMn(II), and Co(II) metal ion in pantadentatemanner and formed stable
complexes. On the basis of IR spectral study it was concluded that ligand coordinated to
metal ion in pantadentate manner i.e. through N2S3donor atoms. IR, UV, EPR spectroscopic
characterization, and molecular modelling study revealed that Mn(II) and Co(II)
complexespossessed an octahedral geometry Fig(8). Molecular modeling calculation results
hold a good comparisonbetween the theoretically predicted geometries and theexperimental
ones.In this study, the compounds were active against testedfungi. The antifungal screening
of all investigated compound provided information about biological activity of ligand and its
complexes which is important in model compounds study.
ACKNOWLEDGEMENT
The one of the author is thankful to the University Grant Commission (UGC) for financial
assistance, USIC university of Delhi for spectroscopic studies, IIT Bombay for ESR, and the
Head, Microbiology Department, SubhartiVivekananda University, Meerut.
REFERENCES
1. Nair R, Shah A, Baluja S, Chanda S. Synthesis and antibacterial activity of some Schiff
base complexes. J. Serb. Chem. Soc. 2006; 71(7): 733-744.
2. Chandra S, Ruchi, Qanungo K, Sharma SK. New hexadentatemacrocyclic ligand and
[image:17.595.153.443.75.237.2]molecular modelling and antimicrobial studies. SpectrochimActa Part A, 2012; 94:
312-317.
3. Sharma AK, Chandra S. Spectroscopic and mycological studies of Co(II), Ni(II) and
Cu(II) complexes with 4-aminoantipyrine derivative. SpectrochimActa Part A, 2011; 81:
424-430.
4. Jain R, Mishra AP. Microwave synthesis and spectral, thermal and antimicrobialactivities
of some novel transition metal complexeswith tridentate Schiff base ligands. J. Serb.
Chem. Soc, 2012; 77(0): 1-27.
5. Gurumoorthy P, Ravichandran J, Karthikeyan N, Palani P, Rahiman AK. Template
Synthesis of PolyazaMacrocyclicCopper(II) and Nickel(II) Complexes: Spectral
Characterization and Antimicrobial Studies. Bull Korean ChemSoc, 2012; 33(7):
2279-2286.
6. Sharma AK, Chandra S. Complexation of nitrogen and sulphur donor Schiff’s base ligand
to Cr(III) and Ni(II) metal ions: Synthesis, spectroscopic and antipathogenic studies.
SpectrochimActa Part A, 2011; 78: 337-342.
7. Hanan F, El-halima A, Omar MM, Mohamed GG. Synthesis, structural, thermal studies
and biological activity of a tridentate Schiff base ligand and their transition metal
complexes. SpectrochimActa Part A, 2011; 78: 36-44.
8. Chandra S, Gupta LK. Modern spectroscopic and biological approach in the
characterization of a novel 14-membered [N4] macrocyclic ligand and its transition metal
complexes. Trans Met Chem, 2007; 32: 240-245.
9. Mobinikhhaled A, Jabbarpour M,Hamta A. Synthesis of some novel and biologically active Schiff base bearing 1,3,4-thiodiazole moiety under acidic and ptc conditions.J
ChilChemSoc, 2011; 56(3): 812-814.
10.Salimon J, Salih N, Yousif E, Hameed A, Ibraheem H. Synthesis, Characterization and
Biological Activity of Schiff Bases of 2, 5-Dimercapto-1,3,4-thiadiazole. Aus J. Basic
ApplSci, 2010; 4(7): 2016-2021.
11.Salimon J, Salih N, Ibraheem H, Yousif E. Synthesis of
2-N-Salicylidene-5-(substituted)-1,3,4-thiadiazole as Potential Antimicrobial Agents, Asian J Chem,2010;
22(7): 5289-5296.
12.PoppFD , Kirsch W. Synthesis of potential anticancer agents. V. Schiff Bases and Related
13.Jarrahpour A, Khalili D, De Clercq E, Salmi C, Michel Brunel JM. Synthesis,
Antibacterial, Antifungal and Antiviral Activity Evaluation of Some New bis-Schiff
Bases of Isatin and Their Derivatives. Molecules 2007; 12: 1720-1730.
14.Shanmugakala R, Tharmaraj P, Sheela CD Anitha C, Synthesis and studies on
s-triazine-based ligand and its metal complexes. Int J InorgChem, doi:10.1155/2012/301086.
15.Prakash A, Adhikari D, Application of schiff bases and their metal complexes-a review,
Int J ChemTech Res, 2011; 3(4):1891-1896.
16.Gopalakrishnan S, Joseph J. Antifungal activities of copper(II) with
biosensitivemacrocyclicschiff base ligands derived from 4-aminoantipyrine derivatives,
Mycobio, 2009; 37(2): 141-146.
17.Chandra S, Bargujar S, Nirwal R, Qanungo K, Sharma SK. Synthesis, spectral
characterization, molecular modeling, thermal study and biological evaluation of
transition metal complexes of a bidentate Schiff base ligand. SpectrochimActa Part A,
2013; 113: 164-170.
18.Chandra S, Kumar U. Spectral studies of coordination compounds of cobalt(II) with
thiosemicarbazone of heterocyclic ketone. Spectrochim. Acta Part A, 2005; 62: 940-944.
19.Bagihalli GB, Avaji PG, Patil SA, Badami PS. Synthesis, spectral characterization, in
vitro antibacterial, antifungal and cytotoxic activities of Co(II), Ni(II) and Cu(II)
complexes with 1,2,4-triazole Schiff bases, Eur J Med Chem, 2008; 43: 2639-2649.
20.Chandra S, Goel S, Dwivedi SD, Spectroscopic and biological studies on newly
synthesized copper(II) and Ni(II) complexes with p-dimethylaminobamzaldehyde
semicarbazone and p-dimethylaminobamzaldehydethiosemicarbazone. Int J Appl Bio
Pharma Tech, 2012; 3: 149-159.
21.Tyagi M, Chandra S, Synthesis, characterization and biocidal properties of platinum
metal complexes derived from 2,6-diacetylpyridine (bisthiosemicarbazone), Open J
InorgChem, 2012; 2: 41-48.
22.Chandra S, Jain D, Sharma AK, Sharma P. Coordination modes of a schiff base
pentadentate derivative of 4-aminoantipyrine with cobalt(II), nickel(II) and copper(II)
metal ions: synthesis, spectroscopic and antimicrobial studies, Molecules, 2009; 14:174-190.
23.Chandra S, Bargujar S, Nirwal R, Yadav N. Synthesis, spectral characterization and
biological evaluation of copper(II) and nickel(II) complexes with thiosemicarbazones
24.Nakamoto K, Infrared and Raman Spectra of Inorganic and Coordination Compounds,
Wiley Interscience, New York, 1978.
25.Ferraro JR, Low Frequency Vibrations of Inorganic and Coordination Compounds,
Plenum Press, New York, 1971.
26.Tyagi M, Chandra S, Tyagi P. Mn(II) and Cu(II) complexes of a bidentate Schiff’s base
ligand: Spectral, thermal, molecular modelling and mycological studies,
SpectrochimicaActa Part A, 2014; 117: 1-8.
27.Chandra S, Gupta LK, Jain D. Spectroscopic studies on Mn(II), Co(II), Ni(II), and Cu(II)
complexes with N-donor tetradentate (N4) macrocyclic ligand derived from
ethylcinnamate moiety, SpectrochimicaActa Part A, 2004; 60: 2411-2417.
28.Lever ABP, Crystal field spectra, in: Inorganic Electronic Spectroscopy, first ed.,
Elsevier, Amsterdam, 1968.
29.Mishra DK, Mishra AP. Synthesis, structural characterization and biological significance
of some novel schiff base complexes with Co(II), Ni(II), and Cu(II), Int J Pharma Res
Develop, 2011; 3: 24-31.
30.Chandra S, Pundir M. Spectroscopic characterization of chromium(III), manganese(II)
and nickel(II) complexes with a nitrogen donor tetradentate, 12-membered
azamacrocyclic ligand. SpectrochimActa Part A, 2008; 69: 1-7.
31.Chandra S, Tyagi M, Sharma K, Mn(II), Co(II), Ni(II) and Cu(II) complexes of a
tertraazamacrocyclic ligand: synthesis, characterization and biological screening, J Iran
ChemSoc,2009; 6(2): 310-316.
32.Drago RS, Physical Methods in Chemistry, Saunders College Publishing, Orlando, 1977.
33.Chandra S, Verma S, Dev U, Joshi N, Tetraazamacrocyclic complexes: synthesis,
spectraland antimicrobial studies, J CoordChem, 2009; 62: 1327-1335.
34.Gopalakrishnan S, Joseph J, Antifungal activities of copper(II) with
biosensitivemacrocyclicschiff base ligands derived from 4-aminoantipyrine derivatives,
Mycobio, 2009; 37(2): 141-146.
35.Mounika K, Anupama B, Pragathi J, and Gyanakumari C, Synthesis¸ characterization and
biological activity of a schiff base derived from 3-ethoxy salicylaldehyde and 2-amino



![Table 5 Optimized Geometry of Legend and Its Complexes (Bond Length In Angstrom) Paremeters Ligand [Mn(L)Cl]Cl [Co(L)Cl]Cl [Co(L)OAc]OAc [Co(L)OAc]OAc](https://thumb-us.123doks.com/thumbv2/123dok_us/786356.585325/12.595.70.530.403.765/optimized-geometry-legend-complexes-length-angstrom-paremeters-ligand.webp)
![Fig. 6 Optimized structure of ligand and complexes (a) ligand (with symbol and numbering) (b) [Mn(L)Cl]Cl, (c) [Mn(L)OAc]OAc, (d) [Co(L)Cl]Cl, (e) [Co(L)OAc]OAc](https://thumb-us.123doks.com/thumbv2/123dok_us/786356.585325/14.595.74.514.77.450/fig-optimized-structure-ligand-complexes-ligand-symbol-numbering.webp)


