metal-organic papers
Acta Cryst.(2006). E62, m625–m627 doi:10.1107/S1600536806006209 Akitsu and Einaga [Cu(C4H4NO2)2(C5H6N2)2].2H2O
m625
Acta Crystallographica Section EStructure Reports Online
ISSN 1600-5368
trans
-Bis(4-aminopyridine-
j
N
)bis(succinimidato-j
N
)copper(II) dihydrate
Takashiro Akitsu* and Yasuaki Einaga
Department of Chemistry, Faculty of Science and Technology, Keio University, 3-14-1 Hiyoshi, Kohoku-ku, Yokohama 223-8522, Japan
Correspondence e-mail: akitsu@chem.keio.ac.jp
Key indicators
Single-crystal X-ray study T= 297 K
Mean(C–C) = 0.007 A˚ Rfactor = 0.052 wRfactor = 0.158
Data-to-parameter ratio = 16.9
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
Received 17 January 2006 Accepted 20 February 2006
#2006 International Union of Crystallography All rights reserved
The title compound, [Cu(C4H4NO2)2(C5H6N2)2]2H2O, has its
metal atom in a square-planar trans-[CuN4] coordination
environment; the CuIIatom lies on a center of symmetry. Both succinimidate and 4-aminopyridine ligands behave as mono-dentate ligands, which coordinate through deprotonated imidate and pyridine (not amino) N atoms, respectively. The 4-aminopyridine is the first ligand in such CuII complexes containing two different N atoms to take potentially different coordination modes.
Comment
Recently, supramolecular self-assembling metal-organic frameworks of transition-metal complexes have been studied widely (Eddaoudiet al., 2001; Evanset al., 2002; Yaghiet al., 1998). In particular, rational design of crystals as well as molecular structures with anisotropy may be important fundamentals in the production of optical and electronic functional materials. It has been found that CuII complexes
show flexible coordination geometries and electronic states (Hathaway & Billing, 1970). We have reported that the tetragonal Jahn–Teller distortion can be affected by the external temperature or light irradiation for CuIIcomplexes with amine ligands (Akitsu & Einaga, 2003). Succinimidate ligands give rise to square-planar CuN4 complexes, not only
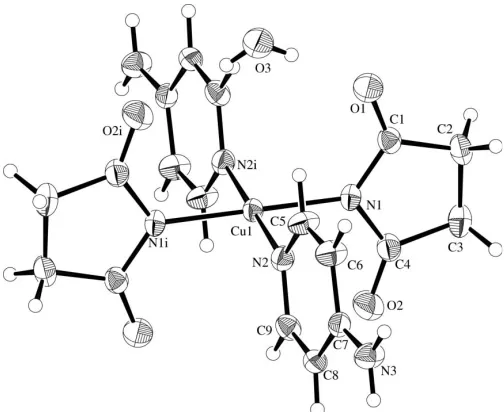
[image:1.610.206.458.507.713.2]with four identical ligands (Akitsu, Komorita & Urushiyama, 2001) but also with mixed ligands such astrans-forms with two succinimidate and two monodentate amine ligands (Akitsu,
Figure 1
Komorita & Kushi, 2001). Furthermore, five-coordinate CuN4O or six-coordinate CuN4O2 square-bipyramidal
chro-mophores have been reported for cis-forms with bidentate diimine (Akitsu et al., 1999) or bidentate amine (Akitsu & Komorita, 2002). However, the related CuII complexes with amine ligands containing two different coordinating N atoms have not been reported so far. Here, we report the crystal structure of the title compound, (I), incorporating 4-amino-pyridine ligands with competitive amino and 4-amino-pyridine N-atom sites.
In (I) (Fig. 1), the Cu atom exists in a square-planar CuN4
coordination geometry; it lies on an inversion center. Both succinimidate and 4-aminopyridine ligands behave as mono-dentate. The Cu—N bond distances, the N—Cu—N bond angles, and other geometric parameters of the ligands (Table 1) are comparable to those of the analogous CuIIcomplexes with pyridine derivative ligands (Latavalya & Taylor, 1975). The C1—N1—C4 angle of 110.2 (4)indicates thesp2
character of the deprotonated N1 atom; the O1—C1—N1 and O2—C4— N1 angles of 123.6 (4) and 124.6 (4), respectively, also reflect
electron delocalization in the-conjugated system, similar to the CuII complexes with 5,5-diphenylhydantoinate ligands (Akitsu & Einaga, 2004, 2005). The dihedral angle between the coordination plane [Cu1/N1/N2/N1i/N2i, symmetry code: (i) x, y, z] and the succinimidate five-membered ring (N1/C1–C4) is 80.2 (2), and that between the coordination
plane and the pyridine ring (N2/C5–C9) is 81.9 (2). The
dihedral angle between the succinimidate and pyridine rings is 85.0 (2).
As seen from Fig. 1, there is a water molecule of crystal-lization (together with an inversion-related one) near the Cu complex to form the O1 H3E—O3—H3F O2i hydrogen-bonding linkage (Table 2). There are also N—H O inter-molecular hydrogen bonds involving the amino groups.
Experimental
Treatment of copper(II) acetate (0.226 g, 1.25 mmol), succinimide (0.495 g, 5.00 mmol), and 4-aminopyridine (0.235 g, 2.50 mmol) in ethanol (50 ml) at 318 K for 2 h gave rise to a deep blue solution. Brown microcrystals (anhydrous) were obtained from the resulting
solution. Yield 0.0752 g (13.2%). IR (KBr): 1618 cm1(amide I). UV (reflectance spectra): 16700 cm1 [sh, F(R
d) = 1.44], 21000 cm
1
[F(Rd) = 2.36]. Anal. Found: C 44.42, H 5.12, N 17.21%; calcd. for C18H24N6CuO6: C 44.67, H 5.00, N 17.36%. m.p. 559 K
(decomposi-tion). Brown crystals of (I) suitable for X-ray analysis were obtained from a methanol solution over a period of several days. The water solvent molecules came from undried methanol.
Crystal data
[Cu(C4H4NO2)2(C5H6N2)2].2H2O
Mr= 483.98 Triclinic,P1 a= 7.945 (2) A˚ b= 8.729 (3) A˚ c= 9.161 (3) A˚ = 65.04 (2)
= 86.86 (3)
= 67.59 (2)
V= 528.2 (3) A˚3
Z= 1
Dx= 1.521 Mg m
3
MoKradiation Cell parameters from 20
reflections = 10.1–13.7
= 1.08 mm1
T= 297 (2) K Prism, brown 0.200.200.20 mm
Data collection
Rigaku AFC-7R diffractometer !–2scans
Absorption correction: scan (Northet al., 1968) Tmin= 0.696,Tmax= 0.807
2736 measured reflections 2430 independent reflections 1771 reflections withI> 2(I)
Rint= 0.058
max= 27.5
h=11!10 k=11!11 l=4!10 3 standard reflections
every 150 reflections intensity decay: 0.4%
Refinement
Refinement onF2
R[F2> 2(F2)] = 0.052 wR(F2) = 0.158
S= 1.06 2430 reflections 144 parameters
H atoms treated by a mixture of independent and constrained refinement
w= 1/[2(F
o2) + (0.0697P)2
+ 0.5544P]
whereP= (Fo2+ 2Fc2)/3
(/)max< 0.001
max= 0.58 e A˚ 3
[image:2.610.123.219.206.359.2]min=0.54 e A˚ 3
Table 1
Selected geometric parameters (A˚ ,).
Cu1—N1 1.971 (3)
Cu1—N2 2.011 (4)
O1—C1 1.235 (6)
O2—C4 1.214 (6)
N1—C1 1.349 (6)
N1—C4 1.371 (6)
N2—C5 1.339 (6)
N2—C9 1.342 (6)
N1—Cu1—N2 89.93 (14)
C1—N1—C4 110.2 (4)
C1—N1—Cu1 127.0 (3)
C4—N1—Cu1 122.8 (3)
C5—N2—C9 116.2 (4)
C5—N2—Cu1 121.5 (3)
[image:2.610.314.565.663.720.2]C9—N2—Cu1 122.0 (3)
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
O3—H3E O1 0.73 (3) 2.13 (2) 2.827 (5) 161 (2)
O3—H3F O2i
0.79 (3) 1.99 (2) 2.770 (7) 168 (2) N3—H3C O1ii
0.86 2.20 3.016 (5) 158
N3—H3D O3iii
0.86 2.08 2.917 (6) 164
Symmetry codes: (i)x;y;z; (ii)xþ1;y1;z; (iii)x;y1;zþ1.
metal-organic papers
The water H atoms were located in difference Fourier maps and their positions were refined withUiso= 0.064 A˚
2
. H atoms bonded to C and N atoms were placed in calculated positions, with C—H and N—H = 0.93–0.97 and 0.86 A˚ , respectively, and with Uiso(H) =
1.2Ueq(C,N).
Data collection: WinAFC Diffractometer Control Software
(Rigaku, 1999); cell refinement: WinAFC Diffractometer Control Software; data reduction:TEXSAN(Molecular Structure Corpora-tion, 1989); program(s) used to solve structure:SIR92(Altomareet al., 1994); program(s) used to refine structure: SHELXL97 (Shel-drick, 1997); molecular graphics:ORTEPII(Johnson, 1976); software used to prepare material for publication:TEXSAN.
This work was supported by a Grant-in-Aid for the 21st Century COE program ‘KEIO Life Conjugate Chemistry’ from the Ministry of Education, Culture, Sports, Science and Technology, Japan. The authors are grateful to Professor Tohru Yamada and Dr Taketo Ikeno (Keio University) for the use of the apparatus for differential scanning calorimetry measurements.
References
Akitsu, T. & Einaga, Y. (2003).Bull. Chem. Soc. Jpn,77, 763–764. Akitsu, T. & Einaga, Y. (2004).Acta Cryst.E60, m524–m526. Akitsu, T. & Einaga, Y. (2005).Acta Cryst.C61, m183–m186. Akitsu, T. & Komorita, S. (2002).Bull. Chem. Soc. Jpn,75, 767–768. Akitsu, T., Komorita, S. & Kushi, Y. (1999).Bull. Chem. Soc. Jpn,72, 447–454. Akitsu, T., Komorita, S, & Kushi, Y. (2001).Inorg. Chim. Acta,315, 18–25. Akitsu, T, Komorita, S. & Urushiyama, A. (2001).Bull. Chem. Soc. Jpn,74,
851–860.
Altomare, A., Cascarano, G., Giacovazzo, C., Guagliardi, A., Burla, M. C., Polidori, G. & Camalli, M. (1994).J. Appl. Cryst.27, 435.
Eddaoudi, M., Moler, D. B., Li, H., Chen, B., Reineke, T. M., O’Keeffe, M. & Yaghi, O. M. (2001).Acc. Chem. Res.34, 319–330.
Evans, O. R. & Lin, W. (2002).Acc. Chem. Res.35, 511–522. Hathaway, B. J. & Billing, D. E. (1970).Coord. Chem. Rev.5, 143–207. Johnson, C. K. (1976).ORTEPII. Report ORNL-5138. Oak Ridge National
Laboratory, Tennessee, USA.
Latavalya, N. & Taylor, M. R. (1975).Cryst. Struct. Commun.4, 163–166. Molecular Structure Corporation (1989).TEXSAN. Version 1.11. MSC, 3200
Research Forest Drive, The Woodlands, TX 77381, USA.
North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968).Acta Cryst.A24, 351– 359.
Rigaku (1999).WinAFC Diffractometer Control Software. Rigaku Corpora-tion, Tokyo, Japan.
Sheldrick, G. M. (1997).SHELXL97. University of Go¨ttingen, Germany. Yaghi, O. M., Li, H., Davis, C., Richardson, D. & Groy, T. L. (1998).Acc. Chem.
Res.31, 474–484.
metal-organic papers
supporting information
sup-1
Acta Cryst. (2006). E62, m625–m627
supporting information
Acta Cryst. (2006). E62, m625–m627 [https://doi.org/10.1107/S1600536806006209]
trans
-Bis(4-aminopyridine-
κ
N
)bis(succinimidato-
κ
N
)copper(II) dihydrate
Takashiro Akitsu and Yasuaki Einaga
trans-Bis(4-aminopyridine-κN)bis(succinimidato-κN)copper(II) dihydrate
Crystal data
[Cu(C4H4NO2)2(C5H6N2)2].2H2O
Mr = 483.98 Triclinic, P1 Hall symbol: -P 1 a = 7.945 (2) Å b = 8.729 (3) Å c = 9.161 (3) Å α = 65.04 (2)° β = 86.86 (3)° γ = 67.59 (2)° V = 528.2 (3) Å3
Z = 1
F(000) = 251.0 Dx = 1.521 Mg m−3
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 20 reflections θ = 10.1–13.7°
µ = 1.08 mm−1
T = 297 K Prism, brown
0.20 × 0.20 × 0.20 mm
Data collection
Rigaku AFC-7R diffractometer
Radiation source: Rigaku rotating anode generator
Graphite monochromator ω–2θ scans
Absorption correction: ψ scan (North et al., 1968)
Tmin = 0.696, Tmax = 0.807
2736 measured reflections
2430 independent reflections 1771 reflections with I > 2σ(I) Rint = 0.058
θmax = 27.5°, θmin = 2.8°
h = −11→10 k = −11→11 l = −4→10
3 standard reflections every 150 reflections intensity decay: 0.4%
Refinement
Refinement on F2
Least-squares matrix: full R[F2 > 2σ(F2)] = 0.052
wR(F2) = 0.158
S = 1.06 2430 reflections 144 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: mixed
H atoms treated by a mixture of independent and constrained refinement
w = 1/[σ2(F
o2) + (0.0697P)2 + 0.5544P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.58 e Å−3
Δρmin = −0.54 e Å−3
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
supporting information
sup-2
Acta Cryst. (2006). E62, m625–m627
O1 0.1355 (5) −0.0913 (5) −0.3077 (5) 0.0609 (9) O2 −0.2430 (6) −0.2366 (6) 0.0641 (5) 0.0742 (12) O3 0.3782 (6) 0.0535 (5) −0.2564 (5) 0.0712 (12) H3E 0.337 (2) −0.005 (3) −0.2640 (6) 0.064* H3F 0.325 (3) 0.107 (3) −0.205 (3) 0.064* N1 −0.0485 (5) −0.1353 (4) −0.1076 (4) 0.0380 (8) N2 0.1873 (5) −0.2370 (5) 0.1653 (4) 0.0394 (8) N3 0.5537 (6) −0.7599 (5) 0.4788 (5) 0.0515 (10) H3C 0.6602 −0.8071 0.4525 0.062* H3D 0.5218 −0.8221 0.5684 0.062* C1 0.0229 (6) −0.1547 (6) −0.2387 (5) 0.0419 (10) C2 −0.0534 (7) −0.2613 (7) −0.2885 (6) 0.0520 (12) H2A −0.1156 −0.1888 −0.3987 0.062* H2B 0.0433 −0.3743 −0.2814 0.062* C3 −0.1862 (7) −0.3016 (7) −0.1694 (7) 0.0547 (12) H3A −0.1545 −0.4329 −0.1114 0.065* H3B −0.3107 −0.2420 −0.2239 0.065* C4 −0.1661 (6) −0.2235 (6) −0.0562 (6) 0.0456 (10) C5 0.3571 (7) −0.3126 (6) 0.1324 (6) 0.0510 (11) H5A 0.3919 −0.2458 0.0355 0.061* C6 0.4815 (6) −0.4820 (6) 0.2333 (6) 0.0505 (11) H6A 0.5978 −0.5272 0.2041 0.061* C7 0.4368 (6) −0.5885 (5) 0.3795 (5) 0.0380 (9) C8 0.2613 (6) −0.5063 (6) 0.4159 (5) 0.0407 (9) H8A 0.2244 −0.5681 0.5138 0.050* C9 0.1447 (6) −0.3364 (6) 0.3084 (5) 0.0431 (10) H9A 0.0285 −0.2861 0.3356 0.052*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
supporting information
sup-3
Acta Cryst. (2006). E62, m625–m627
Geometric parameters (Å, º)
Cu1—N1 1.971 (3) C2—C3 1.503 (7) Cu1—N2 2.011 (4) C2—H2A 0.9700 O1—C1 1.235 (6) C2—H2B 0.9700 O2—C4 1.214 (6) C3—C4 1.505 (6) O3—H3E 0.73 (3) C3—H3A 0.9700 O3—H3F 0.79 (3) C3—H3B 0.9700 N1—C1 1.349 (6) C5—C6 1.360 (6) N1—C4 1.371 (6) C5—H5A 0.9300 N2—C5 1.339 (6) C6—C7 1.390 (6) N2—C9 1.342 (6) C6—H6A 0.9300 N3—C7 1.346 (5) C7—C8 1.399 (6) N3—H3C 0.8600 C8—C9 1.359 (6) N3—H3D 0.8600 C8—H8A 0.9300 C1—C2 1.503 (6) C9—H9A 0.9300
N1—Cu1—N2 89.93 (14) C4—C3—H3A 111.0 H3E—O3—H3F 113 (2) C2—C3—H3B 111.0 C1—N1—C4 110.2 (4) C4—C3—H3B 111.0 C1—N1—Cu1 127.0 (3) H3A—C3—H3B 109.0 C4—N1—Cu1 122.8 (3) O2—C4—N1 124.6 (4) C5—N2—C9 116.2 (4) O2—C4—C3 124.8 (4) C5—N2—Cu1 121.5 (3) N1—C4—C3 110.6 (4) C9—N2—Cu1 122.0 (3) N2—C5—C6 123.5 (4) C7—N3—H3C 120.0 N2—C5—H5A 118.2 C7—N3—H3D 120.0 C6—C5—H5A 118.2 H3C—N3—H3D 120.0 C5—C6—C7 120.8 (4) O1—C1—N1 123.6 (4) C5—C6—H6A 119.6 O1—C1—C2 125.4 (4) C7—C6—H6A 119.6 N1—C1—C2 111.1 (4) N3—C7—C6 122.3 (4) C3—C2—C1 104.2 (4) N3—C7—C8 122.2 (4) C3—C2—H2A 110.9 C6—C7—C8 115.5 (4) C1—C2—H2A 110.9 C9—C8—C7 120.1 (4) C3—C2—H2B 110.9 C9—C8—H8A 119.9 C1—C2—H2B 110.9 C7—C8—H8A 119.9 H2A—C2—H2B 108.9 N2—C9—C8 123.9 (4) C2—C3—C4 103.7 (4) N2—C9—H9A 118.1 C2—C3—H3A 111.0 C8—C9—H9A 118.1
supporting information
sup-4
Acta Cryst. (2006). E62, m625–m627
O1—C1—C2—C3 −178.6 (5) N3—C7—C8—C9 −176.8 (4) N1—C1—C2—C3 1.1 (5) C6—C7—C8—C9 2.5 (6) C1—C2—C3—C4 −3.1 (5) C5—N2—C9—C8 −1.5 (6) C1—N1—C4—O2 176.2 (5) Cu1—N2—C9—C8 172.7 (3) Cu1—N1—C4—O2 −4.4 (7) C7—C8—C9—N2 −0.7 (7) C1—N1—C4—C3 −3.8 (5)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
O3—H3E···O1 0.73 (3) 2.13 (2) 2.827 (5) 161 (2) O3—H3F···O2i 0.79 (3) 1.99 (2) 2.770 (7) 168 (2)
N3—H3C···O1ii 0.86 2.20 3.016 (5) 158
N3—H3D···O3iii 0.86 2.08 2.917 (6) 164