organic papers
Acta Cryst.(2006). E62, o2019–o2020 doi:10.1107/S1600536806013699 Duanet al. C
10H9ClN2O
o2019
Acta Crystallographica Section E Structure Reports Online
ISSN 1600-5368
2-(4-Chlorophenyl)-5-methyl-1
H
-pyrazol-3(2
H
)-one
Xue-Min Duan,a* Mei-Lian Fan,b Peng-Wu Zheng,aJiang-Sheng Lic and Peng-Mian Huangc
aSchool of Pharmacy, Jiangxi Science and
Technology Normal University, Nanchang 330013, People’s Republic of China,bCollege of
Chemistry and Chemical Engineering, Hunan University, Changsha 410082, People’s Republic of China, andcCollege of Pharmaceuticals and Biotechnology, Tianjin University, Tianjin 300072, People’s Republic of China
Correspondence e-mail: dxmlhp@yahoo.com.cn
Key indicators
Single-crystal X-ray study
T= 294 K
Mean(C–C) = 0.003 A˚
Rfactor = 0.038
wRfactor = 0.111
Data-to-parameter ratio = 12.6
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
Received 9 March 2006 Accepted 16 April 2006
#2006 International Union of Crystallography All rights reserved
In the title compound, C10H9ClN2O, the benzene ring is
twisted with respect to the pyrazole plane, with a dihedral angle of 15.81 (11). The centroid-to-centroid separation of 3.721 (4) A˚ indicates–stacking between parallel benzene rings.
Comment
The tautomerism of pyrazolin-5-ones is well known. The crystal structure of the title compound, (I), exhibits the NH tautomer in the crystal state.
The molecular structure of (I) is illustrated in Fig. 1. The pyrazole ring is essentially planar, with a mean deviation of 0.0155 A˚ , but atom H2Ais not coplanar with the ring plane, showing ansp3hybrid nature for atom N2. The dihedral angle between the pyrazole and benzene ring planes is 15.81 (11). The bond lengths and angles are normal (Table 1).
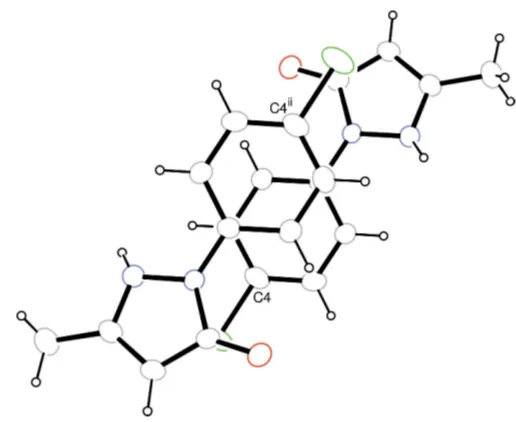
N—H O intermolecular hydrogen bonding occurs in the crystal structure of (I) (Table 2) and – stacking is also observed between parallel benzene rings (Fig. 2), the centroid-to-centroid and face-to-face separations being 3.721 (4) and 3.466 (6) A˚ , respectively.
Experimental
2-(4-Chlorophenyl)-5-methyl-1H-pyrazol-3(2H)-one was prepared according to the literature method of Liu & Li (2004). Single crystals of (I) suitable for X-ray analysis were obtained by evaporation of an ethanol solution.
Crystal data
C10H9ClN2O Mr= 208.64
Triclinic,P1
a= 5.885 (5) A˚
b= 7.704 (7) A˚
c= 11.38 (1) A˚ = 105.980 (13)
= 94.719 (14)
= 101.250 (13)
V= 481.4 (7) A˚3 Z= 2
Dx= 1.439 Mg m 3
Data collection
Bruker SMART CCD area-detector diffractometer
’and!scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin= 0.899,Tmax= 0.925
2397 measured reflections 1669 independent reflections 1421 reflections withI> 2(I)
Rint= 0.025
max= 25.0
Refinement
Refinement onF2 R[F2> 2(F2)] = 0.038 wR(F2) = 0.111
S= 1.07 1669 reflections 132 parameters
H atoms treated by a mixture of independent and constrained refinement
w= 1/[2
(Fo2) + (0.054P)2
+ 0.1893P]
whereP= (Fo2+ 2Fc2)/3
(/)max= 0.003
max= 0.21 e A˚ 3
min=0.26 e A˚ 3
Table 1
Selected bond lengths (A˚ ).
Cl1—C4 1.753 (2) O1—C7 1.256 (2) N1—N2 1.400 (2)
[image:2.610.307.565.67.278.2]N1—C1 1.417 (2) N1—C7 1.392 (2) N2—C9 1.355 (3)
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
N2—H2A O1i
0.84 (3) 1.90 (3) 2.737 (3) 179 (2)
Symmetry code: (i)xþ1;y;z.
Methyl H atoms were placed in calculated psitions, with C—H = 0.96 A˚ , and the torsion angles were refined to fit the electron density. They were treated as riding, withUiso(H) = 1.5Ueq(C). Aromatic H
atoms were positioned geometrically, with C—H = 0.93 A˚ , and they were refined in riding mode, withUiso(H) = 1.2Ueq(C). Atom H2A
was located in a difference Fourier map and refined isotropically. Data collection:SMART(Bruker, 1997); cell refinement:SAINT
(Bruker, 1997); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics:
SHELXTL (Bruker, 1997); software used to prepare material for publication:SHELXTL.
This work was supported by the Foundation of Fine Chemicals Engineering Research Centre for Universities of Jiangxi Province of China.
References
Bruker (1997).SMART,SAINTandSHELXTL. Bruker AXS Inc., Madison, Wisconsin, USA.
Liu, W.-D. & Li, J.-S. (2004).Chin. J. Pestic. Sci.6, 17–21.
Sheldrick, G. M. (1996).SADABS. University of Go¨ttingen, Germany. Sheldrick, G. M. (1997). SHELXS97 and SHELXL97. University of
[image:2.610.44.298.70.202.2]Go¨ttingen, Germany.
Figure 1
The molecular structure of (I), with 30% probability displacement ellipsoids (arbitrary spheres for H atoms).
Figure 2
[image:2.610.44.296.550.583.2]supporting information
sup-1
Acta Cryst. (2006). E62, o2019–o2020
supporting information
Acta Cryst. (2006). E62, o2019–o2020 [https://doi.org/10.1107/S1600536806013699]
2-(4-Chlorophenyl)-5-methyl-1
H
-pyrazol-3(2
H
)-one
Xue-Min Duan, Mei-Lian Fan, Peng-Wu Zheng, Jiang-Sheng Li and Peng-Mian Huang
2-(4-Chlorophenyl)-5-methyl-1H-pyrazol-3(2H)-one
Crystal data
C10H9ClN2O Mr = 208.64 Triclinic, P1 Hall symbol: -P 1 a = 5.885 (5) Å b = 7.704 (7) Å c = 11.38 (1) Å α = 105.980 (13)° β = 94.719 (14)° γ = 101.250 (13)° V = 481.4 (7) Å3
Z = 2 F(000) = 216 Dx = 1.439 Mg m−3 Melting point: 441 K
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 1622 reflections θ = 2.8–26.5°
µ = 0.36 mm−1 T = 294 K Block, colourless 0.30 × 0.26 × 0.22 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
φ and ω scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996) Tmin = 0.899, Tmax = 0.925
2397 measured reflections 1669 independent reflections 1421 reflections with I > 2σ(I) Rint = 0.025
θmax = 25.0°, θmin = 1.9° h = −5→6
k = −9→9 l = −13→10
Refinement
Refinement on F2 Least-squares matrix: full R[F2 > 2σ(F2)] = 0.038 wR(F2) = 0.111 S = 1.07 1669 reflections 132 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H atoms treated by a mixture of independent and constrained refinement
w = 1/[σ2(F
o2) + (0.054P)2 + 0.1893P] where P = (Fo2 + 2Fc2)/3
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
Cl1 0.46931 (12) −0.04642 (9) 0.14776 (5) 0.0653 (3)
O1 0.1306 (2) 0.3563 (2) 0.70412 (13) 0.0514 (4)
N1 0.5211 (2) 0.3831 (2) 0.67330 (13) 0.0340 (4)
N2 0.7367 (3) 0.4918 (2) 0.73944 (15) 0.0397 (4)
C1 0.5052 (3) 0.2847 (2) 0.54670 (16) 0.0324 (4)
C2 0.6975 (3) 0.3122 (3) 0.48463 (18) 0.0414 (5)
H2 0.8339 0.3990 0.5253 0.050*
C3 0.6851 (4) 0.2103 (3) 0.36256 (18) 0.0457 (5)
H3 0.8139 0.2273 0.3214 0.055*
C4 0.4813 (4) 0.0834 (3) 0.30197 (17) 0.0416 (5)
C5 0.2884 (4) 0.0560 (3) 0.36099 (19) 0.0470 (5)
H5 0.1514 −0.0288 0.3188 0.056*
C6 0.2997 (3) 0.1562 (3) 0.48417 (19) 0.0423 (5)
H6 0.1705 0.1377 0.5249 0.051*
C7 0.3443 (3) 0.4248 (3) 0.74289 (17) 0.0362 (4)
C8 0.4626 (3) 0.5550 (3) 0.85669 (17) 0.0418 (5)
H8 0.3916 0.6082 0.9233 0.050*
C9 0.6962 (3) 0.5883 (3) 0.85163 (17) 0.0400 (5)
C10 0.8974 (4) 0.7108 (3) 0.9465 (2) 0.0594 (6)
H10A 0.9931 0.6359 0.9716 0.089*
H10B 0.8384 0.7786 1.0169 0.089*
H10C 0.9895 0.7961 0.9117 0.089*
H2A 0.857 (4) 0.450 (3) 0.730 (2) 0.058 (7)*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
supporting information
sup-3
Acta Cryst. (2006). E62, o2019–o2020
C6 0.0326 (10) 0.0441 (11) 0.0431 (11) 0.0053 (8) 0.0090 (8) 0.0030 (9) C7 0.0287 (10) 0.0431 (10) 0.0367 (10) 0.0115 (8) 0.0095 (7) 0.0081 (8) C8 0.0361 (10) 0.0502 (11) 0.0350 (10) 0.0143 (9) 0.0098 (8) 0.0014 (8) C9 0.0376 (11) 0.0431 (11) 0.0351 (10) 0.0124 (8) 0.0049 (8) 0.0025 (8) C10 0.0396 (12) 0.0672 (15) 0.0509 (13) 0.0088 (10) −0.0002 (9) −0.0110 (11)
Geometric parameters (Å, º)
Cl1—C4 1.753 (2) C3—H3 0.9300
O1—C7 1.256 (2) C4—C5 1.377 (3)
N1—N2 1.400 (2) C5—C6 1.391 (3)
N1—C1 1.417 (2) C5—H5 0.9300
N1—C7 1.392 (2) C6—H6 0.9300
N2—C9 1.355 (3) C7—C8 1.426 (3)
N2—H2A 0.84 (3) C8—C9 1.357 (3)
C1—C2 1.394 (3) C8—H8 0.9300
C1—C6 1.397 (3) C9—C10 1.500 (3)
C2—C3 1.383 (3) C10—H10A 0.9600
C2—H2 0.9300 C10—H10B 0.9600
C3—C4 1.380 (3) C10—H10C 0.9600
C7—N1—N2 108.91 (15) C6—C5—H5 120.2
C7—N1—C1 129.69 (15) C5—C6—C1 119.97 (18)
N2—N1—C1 119.94 (15) C5—C6—H6 120.0
C9—N2—N1 107.48 (16) C1—C6—H6 120.0
C9—N2—H2A 122.4 (17) O1—C7—N1 123.28 (17)
N1—N2—H2A 119.3 (17) O1—C7—C8 131.64 (17)
C2—C1—C6 119.58 (18) N1—C7—C8 105.07 (16)
C2—C1—N1 120.00 (16) C9—C8—C7 108.60 (17)
C6—C1—N1 120.40 (16) C9—C8—H8 125.7
C3—C2—C1 119.96 (18) C7—C8—H8 125.7
C3—C2—H2 120.0 N2—C9—C8 109.75 (17)
C1—C2—H2 120.0 N2—C9—C10 119.95 (19)
C4—C3—C2 119.89 (18) C8—C9—C10 130.30 (19)
C4—C3—H3 120.1 C9—C10—H10A 109.5
C2—C3—H3 120.1 C9—C10—H10B 109.5
C5—C4—C3 121.09 (18) H10A—C10—H10B 109.5
C5—C4—Cl1 119.55 (16) C9—C10—H10C 109.5
C3—C4—Cl1 119.36 (16) H10A—C10—H10C 109.5
C4—C5—C6 119.51 (19) H10B—C10—H10C 109.5
C4—C5—H5 120.2
C7—N1—N2—C9 4.5 (2) C4—C5—C6—C1 0.6 (3)
C1—N1—N2—C9 172.07 (16) C2—C1—C6—C5 0.4 (3)
C7—N1—C1—C2 157.37 (19) N1—C1—C6—C5 −177.99 (17)
N2—N1—C1—C2 −7.2 (3) N2—N1—C7—O1 175.83 (18)
C7—N1—C1—C6 −24.3 (3) C1—N1—C7—O1 9.9 (3)
C6—C1—C2—C3 −1.1 (3) C1—N1—C7—C8 −169.20 (17)
N1—C1—C2—C3 177.26 (17) O1—C7—C8—C9 −178.1 (2)
C1—C2—C3—C4 0.9 (3) N1—C7—C8—C9 0.9 (2)
C2—C3—C4—C5 0.1 (3) N1—N2—C9—C8 −4.0 (2)
C2—C3—C4—Cl1 −179.35 (16) N1—N2—C9—C10 176.84 (19)
C3—C4—C5—C6 −0.8 (3) C7—C8—C9—N2 2.0 (2)
Cl1—C4—C5—C6 178.61 (15) C7—C8—C9—C10 −179.0 (2)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
N2—H2A···O1i 0.84 (3) 1.90 (3) 2.737 (3) 179 (2)