Vol. 60, No. 3 JOURNALOFVIROLOGY,Dec. 1986, p. 1170-1174
0022-538X/86/121170-05$02.00/0
Copyright C 1986, American Society for Microbiology
Three Splicing Patterns Are Used
To
Excise the Small
Intron
Common
to
all Minute
Virus
of Mice RNAs
WILLIAM R. MORGAN1 ANDDAVID C.
WARD'
2*Departments ofHuman Genetics'and Molecular Biophysics and Biochemistry,2 Yale UniversitySchool of Medicine,
New
Haven,
Connecticut 06510Received 25June1986/Accepted 8 September 1986
Weidentified three splicingpatternsusedtoexcise thesmallintroncommontoall threetranscripts encoded
byminute virus of mice. Sequence analysis of minute virus of mice-specific cDNAs indicatedthattwodonor and two acceptorsplice siteswereused:inpattern 1, themostfrequent, nucleotide2280wassplicedtonucleotide
2377; inpattern2, nucleotides 2317and2399werejoined. Oligonucleotideprobes, each specific foroneof the four possible splice junction sequences, were synthesized and hybridized to viral mRNAs immobilized on
nitrocellulosefilters.The probesspecific forsplicepatterns1 and 2 hybridizedtoall three viral mRNAs,asdid a third oligomerspecific for asplicing pattern inwhich nucleotides 2280 and 2399werejoined. The fourth
potential splicingpattern, linking nucleotides 2317 and 2377,wasnotdetected. Thepresenceof three splicing patternsin thetranscripts designated R2 and R3 would allow thetranslationoffive distinct polypeptides from these twomRNAs.
Minute virus of mice (MVM),anautonomousparvovirus,
possesses a linear, single-stranded DNA genome 5,149
nu-cleotides long (1, 2). The transcriptional and translational organization ofMVMDNAhas been elucidated from
exten-sive studies withthe prototypestrain, MVM(p)(Fig. 1). The
three viral mRNAs, transcribed fromtwooverlapping
tran-scription units, are polyadenylated and spliced, and each
includes a small intron between map units 46 and 48 (16).
Although previous experiments have assigned specific viral polypeptide products to each mRNA (5, 6), neither the
absolute number nor the amino acid sequence of these proteins has been established definitively because of
uncer-tainty about the precise splice junctionsand thetranslational initiation sites used.
The initialnucleaseprotection
experiments
usedforRNAmapping indicated that the length ofthe leader exon from
map units 40 to 46 was heterogeneous (16). Furthermore, analysis of the sequence around the small intron identified
numerous potential donor and acceptor splice sites which
could generate nine possible
splicing
patterns (2). Compar-ative sequence studies demonstrated that several autono-mousparvoviruseshadconservedtwoofthepotential5'andone of the potential 3' splice sites in this region of their
genomes (1, 3). In MVM DNA, these sequences occur at
nucleotides 2280, 2317, and 2399, respectively. It has been
hypothesized that the alternativeuse ofthe two conserved
donor sites withthe conservedacceptor site couldgenerate twoforms of themajor viraltranscript R3, which wouldthen code forcapsid protein VP-1 or VP-2. Alternative splicing
within the R3 transcript was further suggested by recent studies with an MVM-bovine papillomavirus chimera
con-taining onlythe downstream (P-39)promoter and therefore presumably transcribing only the R3 mRNA species. Cells
transformed by the chimera DNA expressed both capsid proteins inthe same molarratio as did cells lytically infected with MVM (8).
To establish unambiguously the number and location of splice sites within the MVM transcripts, we deduced viral RNAsequencesfromcDNAs synthesizedbyprimer
exten-*Correspondingauthor.
sion ofaradiolabeled complementary oligonucleotide. The cDNAs were analyzed directly because the overlapping
transcriptional organization complicates identification ofthe RNA origin ofa cDNA and because this approach avoids
potentialartifactsthat could ariseduringtheconstruction or
analysis ofcDNA clones.
Poly(A)+ RNA was isolated from uninfected or
MVM(p)-infected A9 cells 20 to 24 h postinfection by standard methods (11). Oligonucleotides were synthesized on an
Applied BiosystemsDNA synthesizer andpurified by poly-acrylamide-urea gel electrophoresis and then on aSep-Pak column (10). Oligonucleotides were end labeled with
poly-nucleotidekinase(12).Anoligonucleotide complementaryto
viralRNAjust downstream ofthe small intron shared byall viral messages wasusedas aprimer (Fig. 2B). This primer
was hybridized to mRNA from MVM(p)-infected or
uninfectedA9 cells and extendedwithreversetranscriptase essentially asdescribedby Ghoshetal. (7), exceptthatthe
purification of primermRNAhybrids by oligo(dT) cellulose chromatography wasomitted. SeveralspecificcDNAs were
observed with mRNA fromMVM(p)-infected cells
(Fig.
3):twoproducts slightly larger than400nucleotides(B1andB2)
andanother doubletofapproximately0.7kilobases
(Al
andA2).The otherminorbandsobservedwereprobably
prema-turely terminatedcDNAs or derivedfromdegradedRNA. The cDNAs ofapproximately 400 nucleotides were indi-vidually eluted and subjected to Maxam-Gilbert sequence
analysis (12). A different splice junction was identified in each cDNA. In the shorter, more abundant cDNA (B2), nucleotide 2280 (Dl in Fig. 2A) was spliced to nucleotide
2377 (Al, pattern 1); nucleotides 2317 (D2 in Fig. 2A) and 2399(A2,pattern2)werejoinedin the lessabundant, longer
cDNA (B1). Other cDNAs were not
analyzed
further be-causeof insufficientradioactivity.Based on previous nuclease mapping studies (16), full-length primer extension cDNAs derived from intact R3
transcriptswere expectedtobe about 400 nucleotides long,
R2 derivatives were expected to be
approximately
700 nucleotides long, and Ri derivatives were expected to be more than 2kilobaseslong. Althoughweexpectedthat both400-nucleotide-long
cDNAs were derived fromR3,
they
1170
on November 10, 2019 by guest
http://jvi.asm.org/
0 10 20 30 40 50 60 70 80 90 104
TRANSCRIPT
RI - A
MAP
0 UNITS GENE PRODUCT:
NS-I
NS-2
VP-i,
VP-2
m1 mD2 AllA241 PRIMER
NUCLEOTIDE 2250 2300 2350 2400 2450 2500 2550
PATTERN 1 D. Al
PATTERN 2 D2 A2
PATTERN 3
PATTERN 4 -;.SM
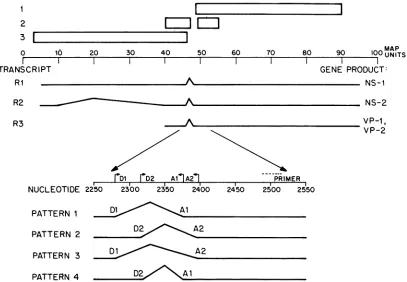
FIG. 1. Transcriptionalandtrahslationalorganizationof the MVMgenome.Themapcoordinates andgeneral splicing patternsof thethree
viraltranscripts, Ri, R2,andR3,asdetermined byPinteletal. (16)areshown inrelationshiptothetwolargeandtwosmallopenreading frames previouslyidentified(2).Thetranslationproductsof eachtranscriptweredeterminedby hybridselectionexperiments (5, 6)andby transfection studies of C127 cells with MVM-bovine papillomaviruschimeras(8).The location of theprimerused for cDNAsynthesisand
thedonor-acceptorsites shown heretogeneratethe alternatesplicingpatternsfor excisionof thecommonintron betweenmapunits 46and
48areindicatedatthe bottom of thefigure.
A
2251 ..-DI
D2
ACTTCAGCGAGCCGCTGAACTTGGACTAA TACGATGGCGCCTCCAGCTAAAAGAGCTAAAAGAGAAGGGTTTAAGGGATGGTTGGTTGGTGGGGTA
TGAAGTCGCTCGGCGACTTGAACCTGATTC-ATGCTACCGCGGAGGTCGATTTTCTCGATTTTCTqCATTCCCAAATTCCCTACCAACCAACCACCCCAT
MetAlaProProAlaLysArgAlaLysArgG
Al A2
TTAATGTTTAATTACCTGT+TTACAGCC+GAAATCACT+GGTTTTA TTGGGTGCCTCCTGGCTACAAGTACCTGGGACCAGGGAACAGCCTTGACCA
AATTACAAATTAATGGACAAAATGT CGGACTTTAGTGAACCAAAT C//t5G GACCGATGTTCATGGACCCTGGTCCCTTGTCGGAACTGGT
yTrpVa ro roGlyTyrLysTyrLeuGlyProGlyAsnSerLeuAspGl
2451
AGGAGAACCAACCAATCCATCTGACGCCGCTGCCAAAGAGCACGACGAGGCCTATGATCAATACATCAAATCTGGAAAAAATCCTTACCTGTACTTCTCy TCCTC'TTGGTTGGTTAGGTAGACTGCGGCGACGGTTTCTCGTGCTGCTC,CGGATACTAGTTATGTAGTTTAGACCTTTTTAGGAATGGACATGAAGAGA
nGlyGluPr oThrAshProSerAspAlaAlaAlaLysGluHisAspGluAlaTy rAspGlnTyrI1eLysSerGlyLysAsnPr oTyrLeuTyrPheSer
2350
2450
2550
B
OLIGONUCLEOTIDE PRIMER
OLIGO A
OLIGO B
OLIGO C
SEQUENCE SPLICE JUNCTION PROBE FOR:
CTCGTGCTGCTCCGGATACTAGTTATGTAG TGAACCTGATTCCGGACTTTAGTG XXXXXXXXXXXX++++++++++++
TCTCGATTTTCTCCMCACCGA
***** *
*******~7
TGAACCTGATTCCAACCCACGGAG
XXXXXXXXXXXX////////////
OLIGO D
Dl TO Al
D2 TO A2
Dl TO A2
[image:2.612.100.507.50.332.2]D2 TO Al
FIG. 2. (A)NucleotidesequenceofMVM(p)neartheregionofsmallintron. Thenucleotidesequenceisrevised from that ofAstellet al. (1). Dl and Al indicate the donor andacceptorsplice sites ofsplicingpattern1,respectively; while D2 and A2similarlyindicate thedonor and acceptorsplice sites of minor splicingpattern 2. Below the nucleotide sequence is theputative amino acidsequence forthe amino terminus ofVP-1(seethetext).Nucleotidecomponentsof thegenomewhicharejuxtaposedintheoligonucleotidesused in these studiesare
indicated by X, *, +, I, or -. Note thatsequences complementarytothe splicejunction-specific oligonucleotides are interruptedin the genomicDNAbyintronsequences.(B)Sequencesofsynthetic oligonucleotide probes.Alloligonucleotidesarecomplementarytoviral RNA. Sequencesarewritteninthe 3'to5' direction foreasyalignmentwithpanelA. Symbolsare asinpanelA.
1171 2
3
F
R3 1
R2
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.612.77.521.422.643.2]1172 NOTES
1632:
A
5 1
.,
..
396 :...:..
FIG. 3. MVM-specific cDNAs migrating as doublets. cDNAs were synthesized by using poly(A)+ RNA from MVM(p)-infected
A9cells. Samples were electrophoresed on a5% polyacrylamide-urea gel until the xylene cyanol migrated about 60 cm (two gel lengths). Lane M, MarkerDNAfragments with nucleotide lengths indicatedattheleft; lane C,cDNAproducts from MVM(p)-infected cells. Al and A2, Minor cDNA bands of about 700 nucleotides; B1 andB2,majorcDNAproductsslightly larger than400nucleotides. The asterisk indicates a cDNA band equivalent in size to that expectedfrom RNA with splicingpattern3(see thetext).
might also have been derived from Rl orR2, either being prematurely terminated products or degraded viral RNA
fragments. To verify that R3 did indeed use both splicing
patternsandtodetermineifthesesamespliceswerepresent as well in
Ri
or R2 transcripts, we probed Northern blotswitholigonucleotides (Fig.
2B)
specific foreachsplicejunc-tion. RNA and DNA samples were
electrophoresed
onagarose-formaldehyde gels and transferred tonitrocellulose
after a mildalkaline treatment by standard procedures (11). The filters were then hybridized with
32P-labeled
MVMplasmidDNA(nicktranslated asdescribed byManiatis et al.
[11]) or with oligonucleotide probes (5' end labeled with
polynucleotide kinase). Posthybridizationwashes weredone as described by Leary et al. (9), except as noted below. Previous studies have shown thatsequences diverging bya
single nucleotide can be distinguished by oligonucleotide probes(4). For
mnaximum
specificity,oligomers complemen-tary to sequences symmetrically spanning eachsplicejunc-tion were used. Under
high-stringency
conditions,
theseoligonucleotide probes should hybridize uniquely to the
appropriate splicejunction and not to unspliced or other,
alternatively splicedmessages. Asexpected,the
oligonucle-otide primer used for cDNA synthesis hybridized to all three viral RNAs,aswellas to anMVM genomicDNA clone(Fig.
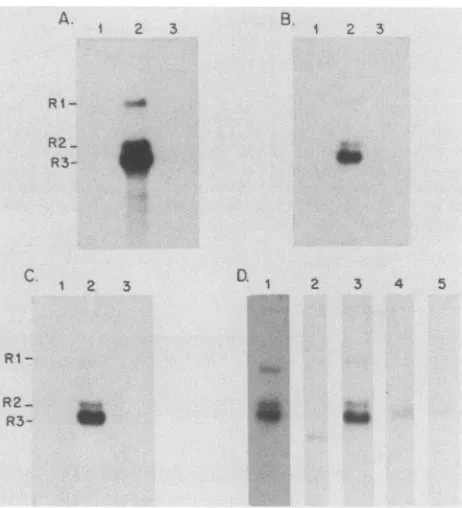
4D, lanes 1 and 2). Similar results were obtained when plasmid pMM984, an infectious MVM clone constructed in pBR322 (14), was used as the probe (data not shown). Interestingly, both the oligomer specific for splicingpattern
1 (oligo A, Fig. 2) and the oligomer specific for splicing pattern 2 (oligo B, Fig. 2) also hybridized to all three viral RNAs with high specificity (Fig. 4A and B, lane 2). In
contrast, no hybridization signal was detected with MVM genomic DNA (Fig. 4A and B, lane 1), a plus-strand genomic RNA synthesized by using SP6 RNA polymerase as de-scribed in Melton et al. (13), or poly(A)+ RNA from uninfected cells (Fig. 4A and B, lane 3) when either the oligo AoroligoBprobewasused, thus confirming the specificity of the splice junction probes. In addition, the ratio of the signals detected byequivalently labeled splice junction oligo A and B probes wassimilar to that seen for the major and
A. 1 2 3 B. 1 2 3
RI- s_
R2 _
R3-C. D. 1
[image:3.612.331.562.272.526.2].. ..*.
...
::
...;
.... ...
...
**kS
...
...k
.. _..
F'.:
...
:.
1 2 3
RI
-
R2-
R3-2 3 4 5
b.
FIG. 4. Oligonucleotide probes specific for each splice junction
hybridizing
toallthreeviralRNAs.Sampleswere electrophoresed onan0.8%formaldehyde-agarose gel, transferredtonitrocellulose filters, and probedwith theoligonucleotides shown in Fig. 2B. In panels A, B, and C, oligo A, oligo B, and oligo C, respectively,werehybridizedtothegenomicDNAclonepMM984 digested with EcoRI (lanes 1), 1 ,ugofpoly(A)+ RNA fromMVM(p)-infected A9 cells (lanes 2), or1 ,ugofpoly(A)+RNAfromuninfectedA9 cells(lanes 3). The specific activity of each oligonucleotide was -2.5 x 108 cpm/,Ig.Autoradiographywasfor18 hat-70°Cwithanintensifying
screen.Inpanel D,each lane wassimilarly exposedtodemonstrate therelativeabundanceofeachsplicingpattern.Lanes1,3, 4,and 5 contain 1 jigofpoly(A)+ RNAfrom MVM(p)-infected cells, and lane2contains pMM984digestedwithEcoRI.Lanes 1 and2were
hybridized with the primer (Fig. 2B), and lanes 3, 4, and5 were
hybridized with oligo A, oligo B, and oligo C, respectively.
Oligo-nucleotidesweresimilarly labeled(-2.5 x 108cpm/,Lg), and
auto-radiographywasfor18 h at roomtemperature withnointensifying
screens. Hybridization was at 42°C for the primer and at room
temperatureforoligo A,oligo B, and oligo C.Stringent washeswere
done in0.2x SSC(lx SSC is 0.15 MNaClplus0.015 M sodium
citrate)at45°Cforpanels A, B, and Dandat40°CforpanelC. J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
NOTES 1173
minor cDNAs
approximately
400 nucleotides long. Thesignal
from each viral RNAwasproportionalto therelative abundance of that RNAspeciesinthe infected cell (16), and themelting
temperatures of the oligomer-RNA hybridswereequivalent
for all three RNAs with each splice junctionprobe (data
notshown). Furthermore, since both oligomerswereradiolabeledtothe samespecific activity, it isapparent that each
transcript
possesses both alternative splicing pat-terns inapproximately
the samerelative molar ratio.Oligonucleotides specific
for the other possible splicing patterns thatusetheidentified
donor and acceptor sites werealso
synthesized
and usedtoprobe
Northernblots. OligoC,which is
specific
for the Dl-A2 splicing pattern (pattern 3),hybridized
toallthree viral RNAs(Fig.
4C), while nosignalwas detected with oligo D, which is specific for the D2-Al
splicing
pattern(data
notshown).
Thehybridization
signalobservedwith
oligo
C wasslightly
lowerthan that seen witholigo
B whenprobes
of identical specific activity and film exposure timeswereused(Fig.
4D, lanes 4 and5), and bothgaveamuch weaker
signal
than didoligoA(Fig.4D, lane 3). A cDNA withsplicing
pattern 3 was probably overlookedduring
ourinitial sequence analysis because of its relatively low abundance.Interestingly,
reinspection of the primer extension data indicated that the band marked with anasterisk in
Fig.
3 had the size expected for a full-lengthcDNA with apattern 3
splice
junction.Based onthe
putative splice
sites conserved among sev-eral autonomousparvoviruses,
previous authors (1, 3)hy-pothesized
thatby alternatively joining
two donor splicesites at nucleotides 2280
(Di)
and 2317 (D2) to a common acceptor siteat2399 (A2), one couldproduce twoforms ofR3 to
differentially
code for VP-2 and VP-1, respectively. Our studies demonstrate that thesetwosplicing
patternsareindeed
used; however,
the most frequently used splicingpattern in MVM
joins
nucleotides2280(Di)
and2377(Al),asplice
sitenotpreviously
identified tobeconserved. Aclosereexamination of the sequences in this region reveals that
feline
panleukopenia
virus(3),H-1 virus (18), canineparvo-virus
(17),
andMVM(i)
(the lymphotropic variant [1])have all conserved an AG dinucleotide approximately 20 basepairs
upstream of the conserved acceptor site previouslyidentified.
Itappears then that ina subset ofR3 transcripts which codes for VP-1, the removal ofnucleotides 2318 to 2398(D2-A2,
pattern
2) allows the AUG atnucleotide2286tobe
spliced
in-frame withthelong right-handopen readingframe
(Fig. 2A).
However, in the majority ofR3 transcripts theputative
VP-1 initiation codon is removed by splicingnucleotide 2280
(Di)
to nucleotide 2377(Al, pattern
1) or 2399(A2,
pattern
3). As previously suggested (3), the re-movalofthisAUGmaybe necessary for efficienttranslationofVP-2
initiating
atnucleotide 2795 (8, 15).Recent
experiments
indicate that the NS-2 protein istranslatedfrom theR2 transcriptand thatreading frame 2 is
used in the second exon, which is just upstream of the
commonintron
(6).
Because translation ofthe second exonoccursin
reading
frame2,
the three splicingpatterns could result in three NS-2 polypeptides with different carboxy termini. Thepredominant
form, translated from messages withsplicing
pattern
1, would have acarboxy terminus of-Leu-Arg-Pro-Glu-Ile-Thr-Trp-Phe.
The two minor species would terminate with -Leu-Arg-Tyr-Asp-Gly-Ala-Ser-Ser(from
pattern
2 RNA) or-Leu-Arg-Leu-Gly-Ala-Ser-Trp-Leu-Gln-Val-Pro-Gly-Thr-Arg-Glu-Gln-Pro
(from pattern 3RNA).
The role that three differentcarboxy peptides mightplay
in thefunction of NS-2is not known.Translation of theNS-1
protein
from theRi
transcripts by use of the longleft-handopen
reading
frame 3 isnotaffectedby
thedifferentsplicing
patterns,since
thisreading
frameterminates
up-streamof both donor sites.Afourth possible pattern, D2-Al, is not used to remove
the common intron.
Experiments
with truncated introns of the rabbit,B-globin
gene (20) showed that efficientsplicing
occurred withanintron of81nucleotides butnotwithoneof
69nucleotides.Indeed,thevastmajority of intronsare
larger
than 75 nucleotides (although the splicing ofshorterintronshas been seen at low efficiency), suggesting that the
mini-mumintron sizefor efficient
splicing
is about 80nucleotides
(19, 20). It is interesting therefore to note that three of the observedsplicing
patterns remove intronslarger
than 80 nucleotides (pattern 1, 96 nucleotides; pattern 2, 81 nucleo-tides; pattern 3, 118nucleotides),
while the fourthsplicing
pattern wouldremove introns of
only
59 nucleotides.This work was supported by Public Health Service grants CA-16038 and AI-19973from the National Institutes of Health.
LITERATURE CITED
1. Astell, C. R., E. M. Gardiner, and P. Tattersall. 1986. DNA sequence of thelymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the
fibrotropicprototype strain. J. Virol. 57:656-669.
2. Astell, C. R., M. Thomas, M. Merchlinsky, and D. C. Ward. 1983. Thecomplete DNA sequence of minute virus of mice,an autonomousparvovirus. Nucleic Acids Res. 11:999-1018. 3. Carlson, J., K. Rushlow, I. Maxwell, F. Maxwell, S. Winston,
and W. Hahn. 1985. Cloning and sequencing of DNAencoding
structural proteins of the autonomous parvovirus feline panleukopeniavirus. J. Virol. 55:574-582.
4. Conner, B. J., A. A. Reyes, C. Morin, K. Itakura, R. L. Teplitz, andR. B.Wallace.1983.Detection of sickle cell 3S-globinallele byhybridizations with synthetic oligonucleotides. Proc. Natl.
Acad. Sci. USA 80:278-282.
5. Cotmore, S. F.,L.J. Sturzenbecker, and P.Tattersall. 1983. The autonomous parvovirus MVM encodes two nonstructural pro-teins in addition to its capsid polypeptides. Virology 129:333-343.
6. Cotmore, S. F., and P. Tattersall. 1986. Organization of nonstructural genes of the autonomous parvovirus minutevirus ofmice. J. Virol.58:724-732.
7. Ghosh, P. K.,V. B. Reddy, M. Piatak,P.Lebowitz,andS. M.
Weissman. 1980. Determination of RNAsequences by primer
directed synthesis and sequencing of their cDNA transcripts.
Methods Enzymol.65:580-595.
8. Labieniec-Pintel, L., and D. Pintel. 1986. The minute virus of mice P39 transcription unit can encode both capsid proteins. J. Virol. 57:1163-1167.
9. Leary, J. J., D. J. Brigati, and D. C. Ward. 1983. Rapid and sensitive colorimetric method for visualizing biotin-labeled DNA probes hybridized to DNA or RNA immobilized on nitrocellulose: bio-blots. Proc. Natl. Acad. Sci. USA 80:4045-4049.
10. Lo, K.-M., S. S. Jones, N. R. Hackett, and H. G. Khorana. 1984. Specific amino acid substitutions in bacterioopsin: replacement of a restriction fragment in the structural gene by synthetic DNA fragments containing altered codons. Proc. Natl. Acad. Sci. USA 81:2285-2289.
11. Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratorymanual, p. 109-112, 191-193, and 202-203. Cold Spring Harbor Laboratory,ColdSpringHarbor,N.Y.
12. Maxam, A. M., andW. Gilbert.1980. Sequencingend-labeled
DNA with base-specificchemicalcleavages.MethodsEnzymol.
65:499-560.
13. Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic VOL.60, 1986
on November 10, 2019 by guest
http://jvi.asm.org/
1174 NOTES
Acids Res. 12:7035-7056.
14. Merchlinsky,M.J., P. J.Tattersall,J.J.Leary,S.F.Cotmore, E. M. Gardiner, and D. C. Ward. 1983. Construction of an
infectious molecular clone of the autonomous parvovirus minutevirus of mice. J.Virol.47;227-232.
15. Paradiso, P. R., K. R.Williams, and R. L. Constantino. 1984. Mapping of the amino terminus of the H-1 parvqvirus major capsid protein. J. Virol. 52:77-81.
16. Pintel, D., D.Dadachanji, C. R.Astell, and D. C. Ward. 1983.
Thegenomeof minute virus of mice,anautonomous
parvovi-rus,encodestwooverlapping transcription units. Nucleic Acids Res. 11:1019-1038.
17. Rhode, S.L., III.1985.Nucleotidesequenceof thecoatprotein
geneof canineparvovirus. J. Virol. 54:630-633.
18. Rhode, S.L.,III,and P. R.Paradiso. 1983. Parvovirusgenome: nucleotidesequenceof H-1 andmapping of itsgenesby hybrid-arrestedtranslation. J.Virol.45:173-184.
19. Upholt, W. B., and L. J. Sandell. 1986.ExonJintronorganization of thechickentypeIIprocollagengene:intronsizedistribution suggests a minimal intron size. Proc. Natl. Acad. Sci. USA 83:2325-2329.
20. Wieringa, B., E. Hofer, and C. Weissmann. 1984. A minimal
intronlength butno specific internal sequenceisrequiredfor
splicingthelargerabbit 1-globinintron. Cell37:915-925.
J.VIROL.