0095-1137/07/$08.00⫹0 doi:10.1128/JCM.01228-06
Copyright © 2007, American Society for Microbiology. All Rights Reserved.
Rapid Multiplex PCR Assay for Identification of USA300
Community-Associated Methicillin-Resistant
Staphylococcus aureus
Isolates
䌤
Kristin K. Bonnstetter,
1Daniel J. Wolter,
1Fred C. Tenover,
2Linda K. McDougal,
2and Richard V. Goering
1*
Department of Medical Microbiology and Immunology, Creighton University School of Medicine, Omaha, Nebraska 68178,1
and Division of Healthcare Quality Promotion, Centers for Disease Control and Prevention, Atlanta, Georgia 303332
Received 15 June 2006/Returned for modification 5 August 2006/Accepted 27 October 2006
Recent reports have noted a discernible increase in the number of community-associated methicillin-resistantStaphylococcus aureus(CA-MRSA) infections in patients without traditional risk factors. In the United States, the most prominent CA-MRSA strain encodes Panton-Valentine leukocidin (PVL) cytotoxin genes, belongs to pulsed field gel electrophoresis type USA300 and multilocus sequence type 8, and carries staphylococcal cassette chromosome mec (SCCmec) type IV. At present, molecular characterization of MRSA strains, such as USA300, can be time-consuming and is often beyond the technical capability of many clinical laboratories, making routine identification difficult. We analyzed the chromosomal regions flanking the SCCmec element in 44 USA300 MRSA isolates and identified a signature “AT repeat” sequence within the conserved hypothetical gene SACOL0058 located 1.4 kb downstream of the 3ⴕend of the J1-SCCmec chromosomal junction. Only USA300 isolates tested contained a sequence of >6 AT repeats in combination with PVL (e.g., related USA500 or Iberian strains had>6 AT repeats but were PVL negative). Using a locked nucleic acid primer specific for>6 AT repeats in combination with primers to detect PVL, we developed a multiplex PCR assay specific for the identification of USA300 strains. Multiplex results were 100% concordant with DNA sequencing, suggesting that the method has promise as a means of rapidly identifying USA300 isolates.
Community-associated methicillin-resistant Staphylococcus
aureus(CA-MRSA) infections are now a major cause of
clin-ical concern. Although first identified in the United States among intravenous drug users (31) followed by other high-risk populations (e.g., prison inmates, athletes, etc.), hospitals na-tionwide have noted an increasing trend in the number of CA-MRSA infections seen in young, healthy populations with-out predisposing risk factors (4, 5, 11, 20, 22, 26). A prospective cohort study published by Naimi et al. found the median age of patients with CA-MRSA infections to be significantly lower than those with healthcare-associated MRSA, at 30 years ver-sus 70 years, respectively (26). Numerous studies have also indicated that CA-MRSA infections are frequently seen among infants and children, again suggesting that the likelihood of con-tracting such infections is no longer limited to traditional at-risk populations (2, 3, 12, 17, 25).
CA-MRSA strains commonly harbor the staphylococcal cas-sette chromosome mec (SCCmec) type IV element and are susceptible to multiple non--lactam antibiotics. This is in con-trast to healthcare-associated strains, such as USA100 isolates, which carry the SCCmectype II element and are resistant to a wide range of antibiotics due to the presence of multiple mo-bile and nonmomo-bile genetic elements (23). However, commu-nity-associated strains typically carry the Panton-Valentine
leukocidin (PVL) geneslukSandlukF, which produce cytotox-ins that cause leukocyte destruction and tissue necrosis (13). Strains producing PVL have been associated with skin abscess formation, furunculosis, and severe cases of necrotizing pneu-monia (21). The presence of PVL genes may also be associated with increased disease severity (6, 10). Despite their commu-nity-associated designation, CA-MRSA strains are frequently isolated from and transmitted among patients within the hos-pital setting (30). CA-MRSA strains have also been associated with increased patient morbidity and mortality, costly treat-ment, and extensive eradication procedures, which under-scores the value of active surveillance for the presence of these strains (29).
Molecular typing methods used to characterize MRSA strains include pulsed-field gel electrophoresis (PFGE), mul-tilocus sequence typing (MLST), and PCR amplification of target genes (32). By PFGE, CA-MRSA isolates in the United States have thus far been classified as pulsed-field types (PFTs) USA300 (sequence type 8 [ST8]), USA400 (ST1) (23), USA1000 (ST59), and USA1100 (ST30) by the Centers for Disease Con-trol and Prevention (L. K. McDougal, W. Zhu, J. B. Patel, and F. C. Tenover, Abstr. 104th Annu. Meet. Am. Soc. Microbiol., abstr. C-220, 2004).S. aureusstrain MW2, responsible for fatal infections in four children from North Dakota and Minnesota between 1997 and 1999, is considered the prototype commu-nity-associated MRSA strain belonging to the USA400 PFT (3). However, recent years have seen an alarming rise in the number of USA300 isolates identified in a variety of commu-nity populations, including children, sports participants, pris-oners, military recruits, and men who have sex with men (1, 2, * Corresponding author. Mailing address: Department of Medical
Microbiology and Immunology, Creighton University School of Med-icine, Omaha, NE 68178. Phone: (402) 280-4098. Fax: (402) 280-1875. E-mail: rgoering@creighton.edu.
䌤Published ahead of print on 8 November 2006.
141
on May 16, 2020 by guest
http://jcm.asm.org/
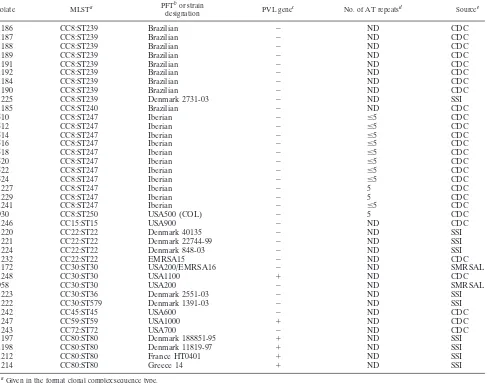
TABLE 1. S. aureusisolate characteristics
Isolate MLSTa PFTbor strain
designation PVL gene
c No. of AT repeatsd Sourcee
956 CC1:ST1 USA400 (MW2) ⫹ ND CDC
CH17 CC1:ST1f USA400 ⫹ ND This study
CH18 CC1:ST1f USA400 ⫹ ND This study
CH33 CC1:ST1f USA400 ⫺ ND This study
CH62 CC1:ST1f USA400 ⫹ ND This study
CH48 CC1:ST1f USA400 ⫹ ND This study
947 CC5:ST5 USA100 (N315) ⫺ 5 CDC
948 CC5:ST5 USA100 (Mu50) ⫺ 5 CDC
1244 CC5:ST5 USA800 ⫺ ⱕ5 CDC
1245 CC5:ST5 USA800 ⫺ ⱕ5 CDC
1226 CC5:ST228 Denmark 3727-03 ⫺ ⱕ5 SSI
1112 CC8:ST8 NCTC8325g ⫺ 5 CDC
1116 CC8:ST8 USA300-0114h ⫹ 6 CDC
1117 CC8:ST8 USA300-0114 ⫹ 6 CDC
1126 CC8:ST8 USA300-0114 ⫹ 6 CDC
1127 CC8:ST8 USA300-0114 ⫹ 6 CDC
1128 CC8:ST8 USA300-0114 ⫹ 6 CDC
1129 CC8:ST8 USA300-0114 ⫹ 8 CDC
1130 CC8:ST8 USA300-0114 ⫹ 8 CDC
1131 CC8:ST8 USA300-0114 ⫹ 6 CDC
1132 CC8:ST8 USA300-0114 ⫹ 8 CDC
1133 CC8:ST8 USA300-0114 ⫹ 6 CDC
1134 CC8:ST8 USA300-0114 ⫹ 6 CDC
1135 CC8:ST8 USA300-0114 ⫹ 6 CDC
1136 CC8:ST8 USA300-0114 ⫹ 6 CDC
1137 CC8:ST8 USA300-0114 ⫹ 6 CDC
1138 CC8:ST8 USA300-0114 ⫹ 6 CDC
1139 CC8:ST8 USA300-0114 ⫹ 6 CDC
1140 CC8:ST8 USA300-0114 ⫹ 8 CDC
1141 CC8:ST8 USA300-0114 ⫹ 6 CDC
1142 CC8:ST8 USA300-0114 ⫹ 6 CDC
1143 CC8:ST8 USA300-0114 ⫹ 6 CDC
1144 CC8:ST8 USA300-0114 ⫹ 6 CDC
1145 CC8:ST8 USA300-0114 ⫹ 8 CDC
1146 CC8:ST8 USA300-0114 ⫹ 6 CDC
1147 CC8:ST8 USA300-0114 ⫹ 6 CDC
1148 CC8:ST8 USA300-0114 ⫹ 6 CDC
1149 CC8:ST8 USA300-0114 ⫹ 6 CDC
1150 CC8:ST8 USA300-0114 ⫹ 6 CDC
1151 CC8:ST8 USA300-0114 ⫹ 6 CDC
1152 CC8:ST8 USA300-0114 ⫹ 6 CDC
1233 CC8:ST8 USA300-0114 ⫹ ⱖ6 CDC
1235 CC8:ST8 USA300-0045i ⫹ ⱖ6 CDC
1335 CC8:ST8f USA300-0045i ⫹ ⱖ6 CDC
1330 CC8:ST8f USA300-0068j ⫹ ⱖ6 CDC
1118 CC8:ST8 USA300-0120k ⫹ 6 CDC
1119 CC8:ST8 USA300-0120 ⫹ 6 CDC
1234 CC8:ST8 USA300-0120 ⫹ ⱖ6 CDC
1236 CC8:ST8 USA300-0120 ⫹ 6 CDC
1336 CC8:ST8f USA300-0120 ⫹ ⱖ6 CDC
1333 CC8:ST8f USA300-0120 ⫹ ⱖ6 CDC
1334 CC8:ST8f USA300-0120 ⫹ ⱖ6 CDC
1337 CC8:ST8j USA300-0120i ⫹ ⱖ6 CDC
1237 CC8:ST8 USA300-0247h ⫹ 6 CDC
1238 CC8:ST8 USA300-0251h ⫹ ⱖ6 CDC
1331 CC8:ST8f USA300-0272l ⫹ ⱖ6 CDC
1228 CC8:ST8 USA500 ⫺ ⱖ6 CDC
1230 CC8:ST8 USA500 ⫺ 6 CDC
1231 CC8:ST8 USA500 ⫺ 6 CDC
511 CC8:ST239 Brazilian ⫺ ND CDC
513 CC8:ST239 Brazilian ⫺ ND CDC
515 CC8:ST239 Brazilian ⫺ ND CDC
517 CC8:ST239 Brazilian ⫺ ND CDC
519 CC8:ST239 Brazilian ⫺ ND CDC
521 CC8:ST239 Brazilian ⫺ ND CDC
523 CC8:ST239 Brazilian ⫺ ND CDC
525 CC8:ST239 Brazilian ⫺ ND CDC
Continued on following page
on May 16, 2020 by guest
http://jcm.asm.org/
15, 18, 24, 27, 28). Detection of USA300 CA-MRSA strains has traditionally required the use of PFGE, MLST, and PCR (i.e., PVL), which, taken together, are time-consuming and require equipment that may not be readily available to the routine clinical laboratory. In addition, the newly described arginine catabolic mobile element recently described by Diep et al. (7) appears to be present only in USA300 strains carrying SCCmec
type IVa (L. K. McDougal, G. E. Fosheim, K. K. Bonnstetter, F. C. Tenover, D. J. Wolter, and R. V. Goering, Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-603, 2006). Thus, the goal of this study was to develop a more
unified molecular approach to the rapid identification of USA300 CA-MRSA isolates.
MATERIALS AND METHODS
Bacterial strains.A total of 106S. aureusstrains (105 MRSA strains and 1
methicillin-susceptibleS. aureusstrain) were examined in this study (Table 1).
[image:3.585.48.545.81.464.2]Strains were chosen according to MLST and PFGE type, with 44 belonging to USA300. Of these, 30 isolates belonged to USA300-0114 clonal complex 8 (CC8: ST8) (9); all were independent isolates known to be epidemiologically unrelated. Other isolates were included on the basis of their genetic relatedness to USA300 strains as determined by MLST BURST analysis (http://www.mlst.net).
TABLE 1—Continued
Isolate MLSTa PFTbor strain
designation PVL gene
c No. of AT repeatsd Sourcee
1186 CC8:ST239 Brazilian ⫺ ND CDC
1187 CC8:ST239 Brazilian ⫺ ND CDC
1188 CC8:ST239 Brazilian ⫺ ND CDC
1189 CC8:ST239 Brazilian ⫺ ND CDC
1191 CC8:ST239 Brazilian ⫺ ND CDC
1192 CC8:ST239 Brazilian ⫺ ND CDC
1184 CC8:ST239 Brazilian ⫺ ND CDC
1190 CC8:ST239 Brazilian ⫺ ND CDC
1225 CC8:ST239 Denmark 2731-03 ⫺ ND SSI
1185 CC8:ST240 Brazilian ⫺ ND CDC
510 CC8:ST247 Iberian ⫺ ⱕ5 CDC
512 CC8:ST247 Iberian ⫺ ⱕ5 CDC
514 CC8:ST247 Iberian ⫺ ⱕ5 CDC
516 CC8:ST247 Iberian ⫺ ⱕ5 CDC
518 CC8:ST247 Iberian ⫺ ⱕ5 CDC
520 CC8:ST247 Iberian ⫺ ⱕ5 CDC
522 CC8:ST247 Iberian ⫺ ⱕ5 CDC
524 CC8:ST247 Iberian ⫺ ⱕ5 CDC
1227 CC8:ST247 Iberian ⫺ 5 CDC
1229 CC8:ST247 Iberian ⫺ 5 CDC
1241 CC8:ST247 Iberian ⫺ ⱕ5 CDC
930 CC8:ST250 USA500 (COL) ⫺ 5 CDC
1246 CC15:ST15 USA900 ⫺ ND CDC
1220 CC22:ST22 Denmark 40135 ⫺ ND SSI
1221 CC22:ST22 Denmark 22744-99 ⫺ ND SSI
1224 CC22:ST22 Denmark 848-03 ⫺ ND SSI
1232 CC22:ST22 EMRSA15 ⫺ ND CDC
1172 CC30:ST30 USA200/EMRSA16 ⫺ ND SMRSAL
1248 CC30:ST30 USA1100 ⫹ ND CDC
958 CC30:ST30 USA200 ⫺ ND SMRSAL
1223 CC30:ST36 Denmark 2551-03 ⫺ ND SSI
1222 CC30:ST579 Denmark 1391-03 ⫺ ND SSI
1242 CC45:ST45 USA600 ⫺ ND CDC
1247 CC59:ST59 USA1000 ⫹ ND CDC
1243 CC72:ST72 USA700 ⫺ ND CDC
1197 CC80:ST80 Denmark 188851-95 ⫹ ND SSI
1198 CC80:ST80 Denmark 11819-97 ⫹ ND SSI
1212 CC80:ST80 France HT0401 ⫹ ND SSI
1214 CC80:ST80 Greece 14 ⫹ ND SSI
a
Given in the format clonal complex:sequence type.
b
Pulsed-field type as described by McDougal et al. (23).
c⫹
, present;⫺, absent.
d
ND, not detected. Numbers indicate the number of AT repeats confirmed with sequencing (numbers withⱕorⱖsigns are the number of AT repeats detected
via PCR assay).
e
CDC, Centers for Disease Control and Prevention, Atlanta, GA; SSI, Robert Skov, Statens Serum Institute, Copenhagen, Denmark; SMRSAL, Donald Morrison, Scottish MRSA Reference Laboratory, Glasgow, United Kingdom.
f
The MLST type has been assumed according to PFGE pattern.
g
This strain is a methicillin-susceptibleStaphylococcus aureusstrain.
h
SCCmectype IVa. USA300-0114 isolates were independent isolates from known geographic locations known to be epidemiologically unrelated.
i
SCCmectype IVb.
j
SCCmectype was nontypeable and was not IVa, IVb, or IVc.
k
USA300-0120 isolates (except isolate 1337) were SCCmectype IVb.
l
SCCmectype IVc.
on May 16, 2020 by guest
http://jcm.asm.org/
PFGE. All strains were analyzed by PFGE. Bacterial DNA was prepared according to the rapid protocol as previously described (14). Pulsed-field pat-terns were analyzed using BioNumerics software (version 4.6; Applied Maths, Kortrijk, Belgium) according to criteria published previously (23).
PCR.Chromosomal DNA was isolated for PCR using the method described by
Enright et al. on the MLST website (http://saureus.mlst.net/misc/info.asp) (8).
Detection of PVL genes was performed using primersluk-PV-1, 5⬘-ATCATT
AGGTAAAATGTCTGGACATGATCCA-3⬘, andluk-PV-2, 5⬘-GCATCAACT
GTATTGGATAGCAAAAGC-3⬘, generating a 433-bp product as described by
Lina et al. (21).
The primers used to detect the conserved hypothetical gene SACOL0058 (S. aureusstrain COL, GenBank accession number CP000046) were ATreg-1,
5⬘-GGAAATGGAATAGAGTTGGCAGAC-3⬘ (nucleotides 69954 to 69977),
and ATreg-2, 5⬘-CAATTAACGATGATATTCCCGATAG-3⬘(nucleotides 70831
to 70855), resulting in an amplification product of 902 bp. Reaction mixtures (100
l total volume) contained 1.5 mM MgCl2, 200M deoxynucleoside
triphos-phate mix, primers at a final concentration of 0.5 mM, 2.5 UTaqDNA
poly-merase (Roche Diagnostics, Mannheim, Germany), and 1l (ca. 1g) of
template DNA. Amplification was carried out for 34 cycles with denaturation at 94°C for 30 s, annealing at 66°C for 30 s, extension at 72°C for 1 min 30 s, and a final extension at 72°C for 5 min.
The primer designed to discriminate the number of AT repeats present within
SACOL0058 was lnaAT: 5⬘-TGL
CTL
CGAL
CGTCAAL
TAL
TATATATAT-3⬘
(nucleotides 69490 to 69511, designed with an additional AT at the 3⬘end of the
primer). Locked nucleic acid (LNA) oligonucleotides (Sigma-Proligo) within the primer are indicated by a superscript L (33). This primer was coupled with the ATreg-2 primer with PCR conditions as described above for detection of
SACOL0058 but with an annealing temperature of 67°C for 30 s and 5 U ofTaq
polymerase, yielding a product 1,366 bp in size.
Multiplex PCR, to simultaneously detect PVL, SACOL0058, and the number of AT repeats, was performed as for SACOL0058, but with primers at the
following concentrations:luk-PV-1,luk-PV-2, and ATreg-1 at 0.05M; ATreg-2
at 0.75M; lnaAT at 0.5M; and 5 U ofTaqDNA polymerase per reaction.
Amplification reactions were visualized by agarose gel electrophoresis (1.5%
SeaKem LE [FMC BioProducts, Rockland, ME]) in 1⫻Tris-borate-EDTA
buffer.
PCR products were sequenced at the Creighton University Molecular Biology Core Facility using an ABI Prism 3100 Avant genetic analyzer (Applied Biosys-tems, Foster City, CA).
RESULTS
Identification of USA300 “signature” AT repeat sequence.
The SCCmecchromosomal region in MRSA isolates is known
to be recombinogenic, resulting in a variety of SCCmectypes (16). We sought potential USA300-specific sequences in genomic regions directly flanking SCCmec, hypothesizing that these areas might also be subjected to higher rates of recom-bination.
[image:4.585.63.520.68.232.2]Using a variety of primers, analysis of ca. 3 kb of genomic sequence upstream of theorfXside of SCCmecrevealed 100% homology across all 30 USA300-0114 (CC8:ST8) strains stud-ied. In addition, the sequence was similar to seven non-USA300Staphylococcus aureusgenomes (MW2, COL, Mu50, N315, NCTC8325, MRSA252, and MSSA476) (http://www.ncbi .nlm.nih.gov and data not shown). However, analysis of 3.3 kb of FIG. 1. Diagram showing the location of the multiplex PCR primers with reference toS. aureusstrain COL (GenBank accession number CP000046). The black bar is the SCCmecresistance element. DR is the 3⬘direct repeat region flanking the SCCmecelement. Gray indicates the 3.3-kb chromosomal sequence 3⬘ to SCCmec, including SACOL0058 and the AT repeat sequence. PCR amplification using primers lnaAT (nucleotides 69490 to 69511) and ATreg-2 (nucleotides 70831 to 70855) indicates the presence ofⱖ6 AT repeats. SACOL0058 is detected using ATreg-1 (nucleotides 69954 to 69977) and ATreg-2. Amplification of the PVL genes using primers designed by Lina et al. (21) occurs at a separate location within the chromosome. The lower black box demonstrates hybridization of the lnaAT primer with the AT repeat sequence as it would occur in a USA300 MRSA strain.
FIG. 2. (A) LNA primer identification of USA300 MRSA strains. Lane 1, 1-kb DNA ladder (Invitrogen, Carlsbad, CA); lanes 2 and 3, USA300:ST8 strains CRG-1130 and CRG-1128, containing eight and six AT repeats, respectively; lane 4, strain CRG-930 (USA500:ST250) containing five AT repeats; and lane 5, water control. (B) Multiplex PCR assay differentiates USA300 strains from other MRSA strains. Lane 1, 1-kb DNA ladder (Invitrogen); lanes 2 and 3, USA300:ST8 strains CRG-1130 and CRG-1128, containing eight and six AT repeats, respectively, as well as SACOL0058 and PVL; lane 4, strain CRG-1112 (mecAnegative) containing five AT repeats; lane 5, strain CRG-1231 (USA500:ST8) containing six AT repeats and SACOL0058; lane 6, strain MW2 (CC1:ST1) (SACOL0058 negative); and lane 7, water control.
on May 16, 2020 by guest
http://jcm.asm.org/
[image:4.585.301.543.477.605.2]genomic sequence extending downstream of the J1-SCCmec
chromosomal junction (19) revealed a region containing either six or eight repetitions of an adenine-thymine base pair in all USA300 isolates examined. Sequence comparison withS. aureus
strain COL (CC8:ST250) located the AT repeat region approxi-mately 1.4 kb downstream from the J1-SCCmec chromosomal junction, within the conserved hypothetical gene SACOL0058. However, SACOL0058 contained only five AT repeats in COL. In addition, PCR analysis revealed the presence of SACOL0058 in PVL-negative USA100, USA500, and USA800 isolates. SACOL0058 was absent from the chromosome of the prototyp-ical community-associated USA400 MRSA strain MW2 (CC1: ST1).
LNA PCR to detect the presence and extent of the AT repeat sequence.Traditional oligonucleotide primers were not suit-able for AT repeat detection due to the potential of the mul-tiple 3⬘ repeats to facilitate in hairpin formation, primer dimers, etc. Therefore, LNA oligonucleotides were used to ensure correct hybridization and discrimination between 5 andⱖ6 AT repeats. This specificity results from the fact that LNA oligonucleotides are modified, with a 2⬘-O, 4⬘-C meth-ylene bridge forming a thermodynamically stable primer with improved target specificity under stringent annealing conditions (33).
An LNA PCR primer was designed with 6 AT repeats at the 3⬘ end and LNA-modified bases near the 5⬘ end to strongly drive correct hybridization and PCR amplification when used with an appropriate reverse primer in isolates with ⱖ6 AT repeats (Fig. 1). As shown in Fig. 2A, amplification was ob-served only in strains containingⱖ6 (i.e., 6 or 8) AT repeats. These results were 100% concordant with DNA sequence anal-ysis (data not shown). LNA PCR was used to examine a variety of strains belonging to MLST clonal complexes CC5 and CC8, including single locus variants of both groups and double or triple locus variants of CC8. In addition to USA300:ST8 iso-lates, SACOL0058 was found in CC8:ST247 (Iberian clone), CC8:ST250 (USA500), CC5:ST5 (USA100 and USA800), and CC5:ST228 isolates but was absent from CC8:ST239 and CC8: ST240, isolates of the Brazilian clone. However, as noted pre-viously, only USA300 isolates, which carry PVL genes, pos-sessed either six or eight AT repeats. Results for all isolates examined are shown in Table 1.
Multiplex PCR for rapid identification of USA300 CA-MRSA isolates.Using minor modification of the PCR, primers were combined to create a multiplex PCR assay with the po-tential to differentiate USA300 isolates from other MRSA strains. As shown in Fig. 2B, lanes 2 and 3, USA300 strains were identified by the amplification of three PCR products: (i) SACOL0058, (ii) LNA-based amplification ofⱖ6 AT repeats, and (iii) PVL genes. A USA500:ST8 isolate, CRG-1231, con-taining six AT repeats, was distinguished from the USA300 isolates by the absence of the PVL band (Fig. 2B, lane 5). The
mecA-negative isolate CRG-1112 (NCTC8325) was a positive control for SACOL0058 (Fig. 2B, lane 4), while strain CRG-956 (USA400, MW2) served as a positive control for PVL (Fig. 2B, lane 6). Discrepant amplification products, when observed, were not problematic due to their minor intensity and size variation.
DISCUSSION
USA300 CA-MRSA isolates are a clear and emerging clin-ical concern. However, the definitive identification of these strains has traditionally involved a combination of tests and protocols (i.e., PFGE, MLST, SCCmec, and PVL) which re-quire specialized expertise and several days to complete. In addition, the arginine catabolic mobile element recently de-scribed by Diep et al. (7) appears to be present only in USA300 strains carrying SCCmec type IVa (L. K. McDougal, G. E. Fosheim, K. K. Bonnstetter, F. C. Tenover, D. J. Wolter, and R. V. Goering, Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-603, 2006). The multiplex assay de-scribed here differentiates USA300 CA-MRSA strains with a variety of SCCmec IV subtypes (Table 1) from other MRSA strains. In this study, only USA300 isolates contained either six or eight AT repeats as well as PVL genes. In some instances, isolates with related sequence types, such as USA500 (ST8), exhibitedⱖ6 AT repeats, but never in combination with PVL. Other isolates (e.g., ST80) encoded PVL but always contained ⬍6 AT repeats. Thus, the combined detection of these ele-ments via multiplex PCR allowed USA300 isolates to be quickly and specifically identified without sequencing. As with any assay, variant strains that could be difficult to detect with this method may exist. Nevertheless, our results suggest that the LNA assay has potential to be used as a rapid, cost-effec-tive approach for identifying USA300 CA-MRSA, a significant pathogen with increasing prevalence in many hospital and community settings.
In the CC8 isolates examined, SACOL0058 was present in ST8, ST247, and ST250 but not in ST239 and ST240, consistent with MLST analysis as discussed by Enright et al. (9). Inter-estingly, SACOL0058 was also found in CC5.
Protein sequence analysis of the conserved hypothetical gene SACOL0058 via the Accelrys GCG translation program (San Diego, CA) showed that MRSA strains containing five AT repeats may possess a fully functional protein. However, an additional AT repeat (i.e., six AT repeats) resulted in a reading frame shift producing a stop codon at amino acid 286 (data not shown). With eight AT repeats, the first 285 amino acids of the protein remain homologous to the original with divergence thereafter. While these data raise serious questions regarding a functional role for SACOL0058 in staphylococcal isolates, the region appears to remain conserved among strains, espe-cially including the USA300 genotype.
AT repeat PCR, in combination with PCR for the presence of PVL genes and SACOL0058, has the potential to identify USA300 CA-MRSA strains in a rapid, cost-efficient manner. Accurate results can be obtained by carefully following opti-mized PCR conditions, allowing valuable diagnostic and sur-veillance data to be collected quickly without the need for sequencing.
ACKNOWLEDGMENT
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
REFERENCES
1.Begier, E. M., K. Frenette, N. L. Barrett, P. Mshar, S. Petit, D. J. Boxrud, K. Watkins-Colwell, S. Wheeler, E. A. Cebelinski, A. Glennen, D. Nguyen, and
on May 16, 2020 by guest
http://jcm.asm.org/
J. L. Hadler.2004. A high-morbidity outbreak of methicillin-resistant Staph-ylococcus aureusamong players on a college football team, facilitated by
cosmetic body shaving and turf burns. Clin. Infect. Dis.39:1446–1453.
2.Buckingham, S. C., L. K. McDougal, L. D. Cathey, K. Comeaux, A. S. Craig, S. K. Fridkin, and F. C. Tenover.2004. Emergence of community-associated
methicillin-resistantStaphylococcus aureusat a Memphis, Tennessee
Chil-dren’s Hospital. Pediatr. Infect. Dis. J.23:619–624.
3.Centers for Disease Control and Prevention.1999. Four pediatric deaths
from community-acquired methicillin-resistantStaphylococcus aureus
—Min-nesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep.48:707–710.
4.Centers for Disease Control and Prevention. 2003. Methicillin-resistant
Staphylococcus aureusinfections among competitive sports participants— Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000-2003.
Morb. Mortal. Wkly. Rep.52:793–795.
5.Centers for Disease Control and Prevention. 2003. Methicillin-resistant
Staphylococcus aureusinfections in correctional facilities—Georgia,
Califor-nia, and Texas, 2001-2003. Morb. Mortal. Wkly. Rep.52:992–996.
6.Chambers, H. F.2005. Community-associated MRSA—resistance and
viru-lence converge. N. Engl. J. Med.352:1485–1487.
7.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington.2006. Complete genome sequence of USA300, an
epidemic clone of community-acquired meticillin-resistant [sic]
Staphylococ-cus aureus. Lancet367:731–739.
8.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt.2000. Multilocus sequence typing for characterization of methicillin-resistant and
methicillin-susceptible clones ofStaphylococcus aureus. J. Clin. Microbiol.
38:1008–1015.
9.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt.2002. The evolutionary history of methicillin-resistant Staphy-lococcus aureus(MRSA). Proc. Natl. Acad. Sci. USA99:7687–7692. 10.Etienne, J.2005. Panton-Valentine leukocidin: a marker of severity for
Staphylococcus aureusinfection? Clin. Infect. Dis.41:591–593.
11.Francis, J. S., M. C. Doherty, U. Lopatin, C. P. Johnston, G. Sinha, T. Ross, M. Cai, N. N. Hansel, T. Perl, J. R. Ticehurst, K. Carroll, D. L. Thomas, E. Nuermberger, and J. G. Bartlett.2005. Severe community-onset pneumonia
in healthy adults caused by methicillin-resistantStaphylococcus aureus
car-rying the Panton-Valentine leukocidin genes. Clin. Infect. Dis.40:100–107.
12.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley.
2005. Methicillin-resistantStaphylococcus aureusdisease in three
communi-ties. N. Engl. J. Med.352:1436–1444.
13.Genestier, A. L., M. C. Michallet, G. Prevost, G. Bellot, L. Chalabreysse, S. Peyrol, F. Thivolet, J. Etienne, G. Lina, F. M. Vallette, F. Vandenesch, and L. Genestier.2005.Staphylococcus aureusPanton-Valentine leukocidin di-rectly targets mitochondria and induces Bax-independent apoptosis of
hu-man neutrophils. J. Clin. Investig.115:3117–3127.
14.Goering, R. V.1993. Pulsed field gel electrophoresis, p. 185–196.InD. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, DC.
15.Gonzalez, B. E., G. Martinez-Aguilar, K. G. Hulten, W. A. Hammerman, J. Coss-Bu, A. Avalos-Mishaan, E. O. Mason, Jr., and S. L. Kaplan.2005. Severe staphylococcal sepsis in adolescents in the era of community-acquired
methicillin-resistantStaphylococcus aureus. Pediatrics115:642–648.
16.Hanssen, A. M., and J. U. Ericson Sollid.2006. SCCmec in staphylococci:
genes on the move. FEMS Immunol. Med. Microbiol.46:8–20.
17.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum.1998.
Community-acquired methicillin-resistantStaphylococcus aureusin children with no
iden-tified predisposing risk. JAMA279:593–598.
18.Hidron, A. I., E. V. Kourbatova, J. S. Halvosa, B. J. Terrell, L. K. McDougal, F. C. Tenover, H. M. Blumberg, and M. D. King.2005. Risk factors for
colonization with methicillin-resistantStaphylococcus aureus(MRSA) in
pa-tients admitted to an urban hospital: emergence of community-associated
MRSA nasal carriage. Clin. Infect. Dis.41:159–166.
19.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu.2003. Insights
on antibiotic resistance ofStaphylococcus aureusfrom its whole genome:
genomic island SCC. Drug Resist. Updates6:41–52.
20.Kazakova, S. V., J. C. Hageman, M. Matava, A. Srinivasan, L. Phelan, B. Garfinkel, T. Boo, S. McAllister, J. Anderson, B. Jensen, D. Dodson, D. Lonsway, L. K. McDougal, M. Arduino, V. J. Fraser, G. Killgore, F. C. Tenover, S. Cody, and D. B. Jernigan.2005. A clone of methicillin-resistant
Staphylococcus aureusamong professional football players. N. Engl. J. Med.
352:468–475.
21.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne.1999. Involvement of Panton-Valentine
leu-kocidin-producing Staphylococcus aureus in primary skin infections and
pneumonia. Clin. Infect. Dis.29:1128–1132.
22.Lindenmayer, J. M., S. Schoenfeld, R. O’Grady, and J. K. Carney.1998.
Methicillin-resistantStaphylococcus aureusin a high school wrestling team
and the surrounding community. Arch. Intern. Med.158:895–899.
23.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover.2003. Pulsed-field gel electrophoresis typing
of oxacillin-resistantStaphylococcus aureusisolates from the United States:
establishing a national database. J. Clin. Microbiol.41:5113–5120.
24.Miller, L. G., F. Perdreau-Remington, G. Rieg, S. Mehdi, J. Perlroth, A. S. Bayer, A. W. Tang, T. O. Phung, and B. Spellberg.2005. Necrotizing fasciitis
caused by community-associated methicillin-resistantStaphylococcus aureus
in Los Angeles. N. Engl. J. Med.352:1445–1453.
25.Mishaan, A. M., E. O. Mason, Jr., G. Martinez-Aguilar, W. Hammerman, J. J. Propst, J. R. Lupski, P. Stankiewicz, S. L. Kaplan, and K. Hulten.2005.
Emergence of a predominant clone of community-acquiredStaphylococcus
aureusamong children in Houston, Texas. Pediatr. Infect. Dis. J.24:201–206. 26.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O’Boyle, R. N. Danila, and R. Lynfield.2003. Comparison of community- and health
care-associated methicillin-resistantStaphylococcus aureusinfection. JAMA290:
2976–2984.
27.Nguyen, D. M., L. Mascola, and E. Brancoft.2005. Recurring
methicillin-resistantStaphylococcus aureusinfections in a football team. Emerg. Infect.
Dis.11:526–532.
28.Pan, E. S., B. A. Diep, E. D. Charlebois, C. Auerswald, H. A. Carleton, G. F. Sensabaugh, and F. Perdreau-Remington.2005. Population dynamics of
nasal strains of methicillin-resistantStaphylococcus aureus—and their
rela-tion to community-associated disease activity. J. Infect. Dis.192:811–818.
29.Rubin, R. J., C. A. Harrington, A. Poon, K. Dietrich, J. A. Greene, and A. Moiduddin.1999. The economic impact ofStaphylococcus aureusinfection in
New York City hospitals. Emerg. Infect. Dis.5:9–17.
30.Saiman, L., M. O’Keefe, P. L. Graham III, F. Wu, B. Said-Salim, B. Kreiswirth, A. LaSala, P. M. Schlievert, and P. Della-Latta.2003. Hospital
transmission of community-acquired methicillin-resistantStaphylococcus
au-reusamong postpartum women. Clin. Infect. Dis.37:1313–1319.
31.Saravolatz, L. D., D. J. Pohlod, and L. M. Arking.1982.
Community-ac-quired methicillin-resistantStaphylococcus aureusinfections: a new source
for nosocomial outbreaks. Ann. Intern. Med.97:325–329.
32.Shopsin, B., and B. N. Kreiswirth.2001. Molecular epidemiology of
methi-cillin-resistantStaphylococcus aureus. Emerg. Infect. Dis.7:323–326.
33.Vester, B., and J. Wengel.2004. LNA (locked nucleic acid): high-affinity
targeting of complementary RNA and DNA. Biochemistry43:13233–13241.