1535-9778/05/$08.00⫹0 doi:10.1128/EC.4.11.1872–1881.2005
Copyright © 2005, American Society for Microbiology. All Rights Reserved.
Comparison of Cell Wall Localization among Pir Family Proteins and
Functional Dissection of the Region Required for Cell Wall
Binding and Bud Scar Recruitment of Pir1p
Toru Sumita, Takehiko Yoko-o, Yoh-ichi Shimma, and Yoshifumi Jigami*
Research Center for Glycoscience, National Institute of Advanced Industrial Science and Technology (AIST), AIST Tsukuba Central 6, Tsukuba, Ibaraki 305-8566, Japan
Received 16 August 2005/Accepted 29 August 2005
We examined the localization of the Pir protein family (Pir1 to Pir4), which is covalently linked to the cell wall in an unknown manner. In contrast to the other Pir proteins, a fusion of Pir1p and monomeric red fluorescent protein distributed in clusters inpir1⌬cells throughout the period of cultivation, indicating that Pir1p is localized in bud scars. Further microscopic analysis revealed that Pir1p is expressed inside the chitin rings of the bud scars. Stepwise deletion of the eight units of the repetitive sequence of Pir1p revealed that one unit is enough for the protein to bind bud scars and that the extent of binding of Pir1p to the cell wall depends on the number of these repetitive units. The localization of a chimeric Pir1p in which the repetitive sequence of Pir1p was replaced with that of Pir4p revealed the functional role of the different protein regions, specifically, that the repetitive sequence is required for binding to the cell wall and that the C-terminal sequence is needed for recruitment to bud scars. This is the first report that bud scars contain proteins like Pir1p as internal components.
Fungal cells are surrounded by a thick cell wall, which is a tough but flexible structure that protects the cell from physical and chemical environmental factors, including pressure, various
shocks, nutrients, and toxins. In the budding yeastSaccharomyces
cerevisiae, the cell wall consists of -1,3-glucan, -1,6-glucan, chitin, and mannose-containing glycoproteins (mannoproteins), together with a small amount of lipids (18). The cell wall proteins (CWPs) are mainly classified into two groups: covalently linked and noncovalently linked to the cell wall (37). The covalently linked cell wall proteins are further classified into glycosylphos-phatidylinositol (GPI)-dependent CWPs (CWPs) and independent alkali-sensitive linkage CWPs (9, 17, 18, 29). GPI-CWPs have a GPI anchor at their carboxyl termini (C termini)
and are immobilized on the cell wall by linking to-1,6-glucan,
which is further coupled to-1,3-glucan (17, 21).
Pir proteins are the main alkali-sensitive linkage CWPs (9). Pir proteins are not modified with a GPI anchor and are directly
bound to-1,3-glucan in the cell wall by an unknown linkage (9,
18). All Pir proteins have one (Pir4p) or several units (Pir1p, Pir2p, and Pir3p) of an internal repetitive sequence. This repeti-tive sequence consists of 18 to 19 amino acid residues at the amino terminus (N terminus) and is the origin of the name Pir
(protein withinternalrepeats) (27, 42). However, the function of
these repetitive sequences is currently unclear.
Genes homologous toS. cerevisiae PIRhave been found in
several yeasts, such asKluyveromyces lactis,Zygosaccharomyces
rouxii(42), Candida albicans(16, 26), andYarrowia lipolytica
(15), but they have not been found in the fission yeast
Schizo-saccharomyces pombe, suggesting thatPIR genes play unique
roles in budding yeasts. Although a lack ofPIRgenes renders
cells sensitive to heat stress (42) and to tobacco osmotin (46), the function of Pir proteins in the cell wall remains unknown.
Functional differences inPIRfamily genes have not been
elu-cidated, although the disruption and expression profile of each
PIRgene has been examined extensively (10, 22, 30, 38, 40, 42).
How the Pir proteins are bound to the cell wall is also unclear. Because Pir proteins can be liberated from the cell wall by mild alkaline extraction and because they are highly O glycosylated, it is thought that they are retained through an
O-glycosidic linkage to-1,3-glucan (9, 19, 29). It has also been
reported that Pir proteins are bound to the cell wall by disul-fide bridges because they can be released by a reducing agent
such as-mercaptoethanol or dithiothreitol (15, 28, 31).
Fur-thermore, Castillo et al. reported that the repetitive sequence of Pir4p is necessary for its binding to the cell wall (5). These results suggest that Pir proteins may be assembled into the cell wall in several different and complex ways.
The purpose of the present study was to elucidate the func-tions of Pir proteins and the mechanism of their translocation to the cell wall. In this report, we show that the distribution of each Pir protein is different, and we focus on the unique lo-calization of Pir1p at the cell wall. Further investigation re-vealed that Pir1p is localized inside the chitin ring of bud scars, which remain at the surface of the mother cell after completion of the budding process. Deletion of the repetitive sequence of Pir1p revealed that the downstream region is actually respon-sible for the recruitment of Pir1p to bud scars. Furthermore, using stepwise deletion, we found that the strength of the linkage between Pir1p and the cell wall depends on the number of repetitive sequence units.
MATERIALS AND METHODS
Strains, growth conditions, and genetic methods.The yeast strains used in this study are listed in Table 1. We usedS. cerevisiaestrainMATatype W303-1A (MATaleu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100; called JHY5) (41) as a wild-type strain.MAT␣type haploid cells (JHY6) were used to make diploid
* Corresponding author. Mailing address: Research Center for Gly-coscience, National Institute of Advanced Industrial Science and Tech-nology (AIST), AIST Tsukuba Central 6, Tsukuba, Ibaraki 305-8566, Japan. Phone: 81-29-861-6160. Fax: 81-29-861-6161. E-mail: jigami .yoshi@aist.go.jp.
1872
on September 8, 2020 by guest
http://ec.asm.org/
cells.PIR1,PIR2,PIR3, andPIR4were disrupted byLEU2,S. pombe his5⫹, TRP1, andURA3, respectively. Thepir1⌬(JTS1),pir2⌬(JTS2),pir3⌬(JTS3), andpir4⌬(JTS4) strains were generated as follows. DNA fragments to disrupt PIR1,PIR2,PIR3, andPIR4were amplified by PCR using primers PIR1-F1/ PIR1-R1, PIR2-F1/PIR2-R1, PIR3-F1/PIR3-R1, and PIR4-F1/PIR4-R1 (Table 2) in combination with LEU2 (J. Horecka, unpublished data), pFA6a-His3MX6, pFA6a-TRP1 (24), and pFA6a-URA3 (J. Horecka, unpublished data) as templates, respectively. The resulting DNA fragments were introduced into W303-1A to create JTS1, JTS2, JTS3, and JTS4, respectively. The compositions of YPD and SD media were as described previously (36), although the YPD medium in the present studies was supplemented with 20g/ml adenine sulfate and is referred to as YPAD medium. YPAD and SD media were used to cultivate yeast cells and to select yeast recombinant transformants, respectively. When necessary, SD medium was supplemented with amino acids, adenine sulfate, and uracil. All yeast cells were grown at 30°C. Yeast transformation was carried out by the lithium acetate method (20).
Escherichia colistrain JM109 {recA1 endA1 gyrA96 thi-1 hsdR17(rK⫺mK⫺)
e14⫺(mcrA⫺)supE44 relA1⌬(lac-proAB) [F⬘traD36 proAB⫹lacIq
lacZ⌬M15]} was used for plasmid preparations and subcloning.
Visualization of Pir proteins and bud scars.To visualize Pir1p, Pir2p, Pir3p, and Pir4p, a gene encoding monomeric red fluorescent protein (mRFP) (4) was fused withPIR1,PIR2,PIR3, andPIR4, respectively. PIR1 andPIR4 open reading frames (ORFs) were fused with the mRFP gene at their 3⬘ends (see Fig. 1A and 6A), andPIR2andPIR3ORFs were fused with the mRFP gene downstream of the site recognized by Kex2p protease (see Fig. 5A and 6A). A BamHI site was created upstream of the stop codon ofPIR1and downstream of the Kex2p cleavage site ofPIR2andPIR3. The mRFP gene, which has a BamHI site both upstream of the start codon and at the end of the ORF without the stop codon, was introduced in frame into the above BamHI sites. Similarly, a SpeI site was created upstream of the stop codon ofPIR4. The mRFP gene, which has a SpeI site both upstream of the start codon and at the end of the ORF without the stop codon, was introduced in frame into the above SpeI site.PIR1-mRFP, mRFP-PIR2, andPIR4-mRFP fusion genes were expressed under the control of their own promoters on pRS304 in JTS1, JTS2, and JTS4, respectively. The mRFP-PIR3fusion gene was expressed under the control ofPIR3promoter on pRS305 in JTS3. The resulting plasmids (pRS304-PIR1-mRFP, pRS304-mRFP-PIR2, pRS305-mRFP-PIR3, and pRS304-PIR4-mRFP) were linearized and in-troduced into JTS1, JTS2, JTS3, and JTS4 to create JTS1-1m, JTS2-m2, JTS3-m3, and JTS4-4m, respectively.
Pir1p-mRFP in diploid cells was expressed as follows. ThePIR1of strain JHY6 was disrupted withS. pombe his5⫹to generate JTS1␣. pRS306 carrying the PIR1-mRFP gene was linearized by cleavage with EcoRV and introduced into JTS1␣for chromosome integration at theURA3locus. The resulting strain was mated with JTS1-1m to generate strain JTSD-1m.
Bud scars were stained with calcofluor white (CFW; Sigma-Aldrich, St. Louis, MO) at a final concentration of 1g/ml for 10 min or with fluorescein isothio-cyanate (FITC)-labeled wheat germ agglutinin (WGA, lectin fromTriticum
vul-garis; Sigma-Aldrich, St. Louis, MO) (FITC-WGA) (32) at a final concentration of 40g/ml for 10 min.
Gene disruption.BNI1,SPA2,PEA2,AXL2,BUD3,BUD8,RGA1,BEM1, BEM2,VMA6,APN1,CHS2, andCHS3in strain JTS1-1m were disrupted with S. pombe his5⫹. The disruption cassette was PCR amplified using synthetic oli-gonucleotides as primers (Table 2) and pFA6a-His3MX6 as a template. For example, theBNI1disruption cassette was PCR amplified using the primers BNI1-F1 and BNI1-R1. Each resulting DNA fragment was introduced into JTS1-1m.
Microscopic observation.Yeast cells were cultivated in 2 ml of YPAD medium until the late exponential phase at 30°C. They were then centrifuged (2,500⫻g, room temperature, 5 min), washed twice, and suspended in 500l of phosphate-buffered saline buffer (10 mM sodium phosphate and 150 mM sodium chloride, pH 7.4). For three-dimensional (3-D) observation, the cells were suspended in 500l of phosphate-buffered saline containing 0.1% agarose.
We used a BX50 fluorescence microscope (Olympus, Tokyo, Japan) and MicroMAX cooled charge-coupled device (CCD) camera (Roper Scientific, Duluth, GA) for observation of staining. For 3-D observation, we used an IX71 fluorescence microscope (Olympus) with a confocal scanner, a piezo-actuated prototype 3-D system (Yokogawa Electric, Tokyo, Japan), and a high-gain ava-lanche rushing amorphous photoconductor camera system (Hitachi Kokusai Electric and NHK, Tokyo, Japan). We also used an EVM285SPD cooled CCD camera (Texas Instruments, Dallas, TX) instead of a high-gain avalanche rushing amorphous photoconductor camera for enlarged images of the bud scars. The photographic images were analyzed using Adobe Photoshop, version 5.0.
Deletion of the repetitive sequence ofPIR1.The repetitive sequence ofPIR1was deleted stepwise by PCR as follows. A BamHI site was created downstream of the Kex2p cleavage site ofPIR1on pUC118 to generate pUC-PIR1B. Several regions of thePIR1ORF were PCR amplified using primers PIR1-A, PIR1-B, PIR1-C, PIR1-D, PIR1-E, and PIR1-F in combination with primer PIR1-2550 (Table 2). Each amplified DNA fragment was digested with BamHI and ClaI and then ligated into the BamHI-ClaI region of pUC-PIR1B. Each resulting plasmid was digested with BglII and ClaI and ligated into the BglII-ClaI region of pRS304-PIR1-mRFP. These plasmids were linearized by EcoRV digestion and introduced into JTS1 for chromosome integration at theTRP1locus. These transformants contain 5, 4, 3, 2, 1, and 0 repetitive sequence units and were named JTS1-A, JTS1-B, JTS1-C, JTS1-D, JTS1-E, and JTS1-F, respectively (see Fig. 7A).
Expression of the Pir4/Pir1 chimeric protein.The repetitive sequence ofPIR4 and the downstream region of the repetitive sequence of PIR1 were fused together. PIR4-RepA and PIR4-RepB (Table 2) were chemically synthesized, annealed, phosphorylated with T4 polynucleotide kinase, and inserted into the BamHI-SmaI site of pBluescript II SK⫹(pBS) to generate pBS-PIR4Rep. The insert of this plasmid contains the repetitive sequence ofPIR4, which has an HpaI site at its 3⬘end. The downstream region of the repetitive sequence ofPIR1 was PCR amplified using primers PIR1-HpaI and M13 with pUC-PIR1B as a template. The amplified fragment was digested with HpaI and HindIII and inserted into the HpaI-HindIII site on pBS-PIR4Rep. The resulting plasmid was
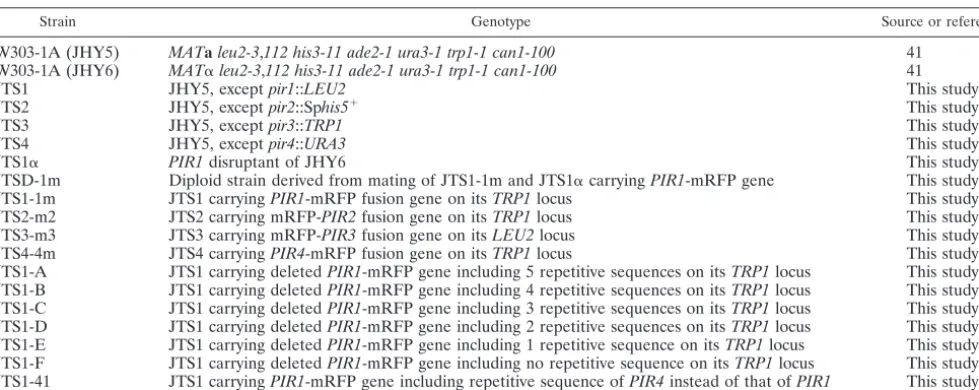
TABLE 1. Yeast strains used in this study
Strain Genotype Source or reference
W303-1A (JHY5) MATaleu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100 41
W303-1A (JHY6) MAT␣leu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100 41
JTS1 JHY5, exceptpir1::LEU2 This study
JTS2 JHY5, exceptpir2::Sphis5⫹ This study
JTS3 JHY5, exceptpir3::TRP1 This study
JTS4 JHY5, exceptpir4::URA3 This study
JTS1␣ PIR1disruptant of JHY6 This study
JTSD-1m Diploid strain derived from mating of JTS1-1m and JTS1␣carryingPIR1-mRFP gene This study
JTS1-1m JTS1 carryingPIR1-mRFP fusion gene on itsTRP1locus This study
JTS2-m2 JTS2 carrying mRFP-PIR2fusion gene on itsTRP1locus This study
JTS3-m3 JTS3 carrying mRFP-PIR3fusion gene on itsLEU2locus This study
JTS4-4m JTS4 carryingPIR4-mRFP fusion gene on itsTRP1locus This study
JTS1-A JTS1 carrying deletedPIR1-mRFP gene including 5 repetitive sequences on itsTRP1locus This study JTS1-B JTS1 carrying deletedPIR1-mRFP gene including 4 repetitive sequences on itsTRP1locus This study JTS1-C JTS1 carrying deletedPIR1-mRFP gene including 3 repetitive sequences on itsTRP1locus This study JTS1-D JTS1 carrying deletedPIR1-mRFP gene including 2 repetitive sequences on itsTRP1locus This study JTS1-E JTS1 carrying deletedPIR1-mRFP gene including 1 repetitive sequence on itsTRP1locus This study JTS1-F JTS1 carrying deletedPIR1-mRFP gene including no repetitive sequence on itsTRP1locus This study JTS1-41 JTS1 carryingPIR1-mRFP gene including repetitive sequence ofPIR4instead of that ofPIR1 This study
on September 8, 2020 by guest
http://ec.asm.org/
digested with BamHI and ClaI, and the resulting fragment containing the repet-itive sequence ofPIR4was ligated into the BamHI-ClaI region on pUC-PIR1B. The plasmid thus generated was digested with BglII and ClaI and then ligated into the BglII-ClaI region ofPIR1-mRFP on pRS304. This chimeric gene was expressed under the control ofPIR1promoter in JTS1 and was referred to as JTS1-41 (see Fig. 9A).
Preparation of Pir1p-mRFP.Pir1p-mRFP fusion proteins were directly pre-pared from whole cells by a mild alkaline treatment. The cells were cultivated overnight in 2 ml of YPAD medium, after which 300l of the suspension was seeded in 15 ml of fresh YPAD medium and further cultivated for 6 h until the cell wet weight was 10 mg/ml. The cells were centrifuged and washed three times. The fusion proteins were extracted by treating the cells overnight at 4°C with 1 ml of 30 mM NaOH. The cells were centrifuged, and proteins in the supernatants were recovered by trichloroacetic acid precipitation. The proteins released into the medium during cultivation were collected overnight at 4°C by adding 111l of 100% (wt/vol) trichloroacetic acid to 1 ml of the medium, followed by cen-trifugation (13,000⫻g, 4°C, 20 min) to precipitate the protein.
Western blotting.Proteins were separated by sodium dodecyl sulfate-poly-acrylamide gel electrophoresis on a 5 to 20% gradient gel (Atto, Tokyo, Japan) and then transferred to a Hybond-P polyvinylidene difluoride membrane (Am-ersham, Piscataway, NJ). Pir1p-mRFP and the truncated derivatives were de-tected with an immunopurified polyclonal antibody against mRFP followed by
horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G (Am-ersham). The polyclonal antibody against mRFP was obtained from TaKaRa Co., Ltd. (Tokyo, Japan). The C-terminal region of the mRFP protein (Glu212-Ala225) was chemically synthesized and coupled to keyhole limpet hemocyanin through the cysteine residue of the peptide. This conjugate was injected into a rabbit to obtain an antipeptide antibody, which was purified by sequential chro-matographic steps on protein A and peptide columns. Immunoreactive bands were visualized using the ECL plus Western blotting detection system (Amer-sham) and with a LAS-1000 luminescence image analyzer (Fuji Photo Film, Tokyo, Japan).
RESULTS
Pir1p localizes inside the chitin rings of bud scars.Although Pir1p had been isolated as a cell wall protein, the exact local-ization of Pir1p at the cell wall had not been reported. We constructed a Pir1p fusion protein tagged with mRFP at the C terminus to monitor the cellular localization (Fig. 1A). We confirmed that Pir1p was expressed on the cell surface. Inter-estingly, Pir1p was visualized as one or several spots on the cell
TABLE 2. Synthetic oligonucleotides used for gene deletion, cloning, and tagging
Oligonucleotide Sequence (5⬘–3⬘)
PIR1-F1...CTAATTATTCTTTCAATAAGAAGATCGTAATGCTACAATTCTCACATATACGGATCCCCGGGTTAATTAA PIR1-R1 ...AATATGCTACGATGACTTTATGTTTTCATGCGACTATGAGAGGTAAACTTGAATTCGAGCTCGTTTAAAC PIR2-F1...TTTGAGCTAAATTTAGATTTTGGTCAAAATAAGAAAGATCCTAAAAAAGGCGGATCCCCGGGTTAATTAA PIR2-R1 ...ATGCAAATGAGAAAAAGTCATCAGGATAAAAGAACTAATAGTTTTCTGCTGAATTCGAGCTCGTTTAAAC PIR3-F1...ATCTCCAGGATTATCCCTACTCCATTCATTACAACATGCGCCAAATCAAGCGGATCCCCGGGTTAATTAA PIR3-R1 ...CAACTAATTCGTGAAGTTAAAGGAGGACGACTCCGATTGATCGATGCATCGAATTCGAGCTCGTTTAAAC PIR4-F1...ACAAAAACAAAAATAAGAAATAGGTTGACAGTGGGTGAAAAATTCTCGAACGGATCCCCGGGTTAATTAA PIR4-R1 ...TTTTCTATTTCTTTTCCCTTATAAAAGGTAGAACATTAGTACTTTGATGTGAATTCGAGCTCGTTTAAAC M13...GTAAAACGACGGCCAGT
PIR1-A ...AAGGGATCCTCAGCTAAGACTACCG PIR1-B ...AAGGGATCCGCTGCAGCCGT PIR1-C ...AAAGGATCCGCTGCTGCTGTC PIR1-D...AAAGGATCCGCTGCAGCTGTCT PIR1-E ...AAAGGATCCGCTGCTGCTGTTTC PIR1-F...AATGGATCCGTTGCTCCAGTCTC PIR1-2550 ...CGACTATGAGAGGTAAAC
PIR4-RepA...GATCCGACGTTATCTCTCAAATTGGTGACGGTCAAGTCCAAGCCACTGTTAACCCC PIR4-RepB ...GGGGTTAACAGTGGCTTGGACTTGACCGTCACCAATTTGAGAGATAACGTCG PIR1-HpaI ...CTTTAGTTAACGCAACCATTATACCATC
BNI1-F1 ...CTGAACCTTTTCAACAAACGAGAAGCAAGAAAAGAAGAGAAGGAAAGGAACGGATCCCCGGGTTAATTAA BNI1-R1...TGGATGTTTGTTTTGGTATTACTGTTGTCATAATTTTTTGGTTTAATATTGAATTCGAGCTCGTTTAAAC SPA2-F1 ...GCAATACGAGCCACCGAAACAGAATAAACAAAAGAAAAGAAAGAGTAAACCGGATCCCCGGGTTAATTAA SPA2-R1 ...TATTGATTGTCTTTGTCTTCCTTTTCTTTCTCCTCTAGATACTACTAAGTGAATTCGAGCTCGTTTAAAC PEA2-F1 ...TCGAAGTCCTGTGTTCGAGAGGGAGAATAACGGTGGATATCACGTTTCATCGGATCCCCGGGTTAATTAA PEA2-R1...TTCTTCTTATTCTATATTTATATATCAATGTTTTATAATAAGATGTTTATGAATTCGAGCTCGTTTAAAC AXL2-F1...ACGATTTCCTGCTTCCAACATCTACGTATATCAAGAAGCATTCACTTACCCGGATCCCCGGGTTAATTAA AXL2-R1 ...TAAAAAATAAAACAGGAAAATAAAATTAAGCAAAATATCGTTGCGTATAAGAATTCGAGCTCGTTTAAAC BUD3-F1 ...GTGTTCGGGCTGTTATCTGGTTGCTAAAAGAGTATATTTACACCTCACCACGGATCCCCGGGTTAATTAA BUD3-R1...AATGTATACATTGCATTAAATTAAAAAGAAAAAAAAAATCAATAAAACACGAATTCGAGCTCGTTTAAAC BUD8-F1 ...TACCCAATATCCTCTTTCTACTTGAGCCTTTTTTCGATTCTACATGAAGTCGGATCCCCGGGTTAATTAA BUD8-R1...GACAGAACAGTTTTTTATTTTTTATCCTATTTGATGAATGATACAGTTTCGAATTCGAGCTCGTTTAAAC RGA1-F1 ...ACAAGGATAGCTGATTCAGGTACTAGTGGTGGAGAGAGCGGCATATTAAACGGATCCCCGGGTTAATTAA RGA1-R1...GTTTCGATACAGTTCATATAAGGCGGCTCAATGCAGAACCGAGGATAGCGGAATTCGAGCTCGTTTAAAC BEM1-F1 ...TGCACGTTGAAAGCACTGTGTGAAAAGAAATTGTCAAGAAAGCCATATAACGGATCCCCGGGTTAATTAA BEM1-R1...GTGCATCTGCCAAGTAAAGAAGAAAAATGCTTCGTCTTCTAACACTAGATGAATTCGAGCTCGTTTAAAC BEM2-F1 ...TGCCTTTTCTGGATAGACACAAAAAAAACAAATAACGAAGCAGGAGTCTACGGATCCCCGGGTTAATTAA BEM2-R1...AATTTTTTCTCTCTCAGCAGTGGATTGTATACATTTACCACGAAAATTGTGAATTCGAGCTCGTTTAAAC VMA6-F1...TCGTTCTTTCATTCTTAGAGTTAAAAAGCAAATAGAGAAGAAAAGAAACACGGATCCCCGGGTTAATTAA VMA6-R1 ...GAAGAAAAGACATCTAACAAATATACCAGAAGATAAATGCTACATATATCGAATTCGAGCTCGTTTAAAC APN1-F1 ...AAACACAAAACGCAACATTAATAAGCTTTTGGCATATCGGAACCATCGTACGGATCCCCGGGTTAATTAA APN1-R1 ...AGATAATCTACAAAAATTGATTACGTATTTAAAATTCTTCTCGCTTCTCAGAATTCGAGCTCGTTTAAAC CHS2-F1 ...GAGGAAGTTACATATAGACCCAAATAAAAACCAAAGAACCACATATAGAACGGATCCCCGGGTTAATTAA CHS2-R1...CTAAAACAGAATGAGAAAAAAGAGGGAATGACGAGAAATTAGCTGAAAAAGAATTCGAGCTCGTTTAAAC CHS3-F1 ...CCATTTTCTTCAAAGGTCCTGTTTAGACTATCCGCAGGAAAGAAATTAGACGGATCCCCGGGTTAATTAA CHS3-R1...CACAACCATATATCAACTTGTAAGTATCACAGTAAAAATATTTTCATACTGAATTCGAGCTCGTTTAAAC
on September 8, 2020 by guest
http://ec.asm.org/
surface. A similar result was obtained when Pir1p was tagged with mRFP at its N terminus (data not shown), indicating that Pir1p localizes in clusters. A more precise observation revealed that some spots of Pir1p take the form of a ring-like structure (Fig. 1B).
Based on this result, we expected that Pir1p is localized at bud scars. Because the bud scar is mainly composed of chitin, we confirmed this by staining the cells with CFW, which spe-cifically binds chitin (Fig. 1B). As expected, the positions of the Pir1p spot and of the bud scar were very similar, clearly indi-cating that Pir1p is localized at bud scars.
To investigate the precise localization of Pir1p around the chitin ring of bud scars, we observed the cells with a confocal microscope connected with cooled CCD camera system and then enlarged the image of the bud scars using digital imaging software. Merging of the chitin ring and Pir1p localization revealed that they did not overlap; the ring structure of Pir1p is obviously located inside the chitin ring (Fig. 2A). We
ob-tained similar results using FITC-WGA (lectin fromTriticum
vulgaris) instead of CFW to stain the chitin ring (Fig. 2B). These results indicate that Pir1p localizes inside the chitin ring of bud scars.
We further observed the chitin ring of bud scars and Pir1p-mRFP stereoscopically by confocal fluorescent microscopy. The 3-D observation indicates not only that the ring structure of Pir1p is located inside the chitin ring but also that not all bud scars contain Pir1p (see the supplemental material at http: //unit.aist.go.jp/rcg/rcg-gb/pir1.html). Because the ring struc-ture of Pir1p-mRFP cannot be observed at the budding site before cell separation is completed, this result suggests that Pir1p localizes inside the chitin ring of bud scars after cell separation has completed.
The budding pattern of diploid cells is different from that of haploid cells (8). To further investigate the localization of
Pir1p, Pir1p-mRFP was expressed in diploid cells.PIR1 was
disrupted in JHY6 (MAT␣ type) cells, and the fusion gene
encoding Pir1p-mRFP was introduced. After mating of this strain and JTS1-1m, we observed the distribution of Pir1p in the resulting diploid. As shown in Fig. 3, Pir1p localized inside the chitin ring of bud scars in diploid cells as expected. A similar result was obtained when the chitin ring was stained with FITC-WGA instead of CFW (data not shown). These findings confirm that Pir1p is expressed at bud scars both in haploid and diploid cells.
Translocation of Pir1p to the budding site is not affected in
chs2⌬andchs3⌬cells.Next, we investigated the relationship between Pir1p and chitin. Before mother and daughter cells separate, chitin synthase III (CSIII) catalyzes the synthesis of the chitin ring at the base of an emerging bud. The primary septum is then formed by CSII, which is the first step in the division of these cells. After the primary septum is completed, secondary septa are laid down from the mother and daughter
FIG. 1. (A)PIR1-mRFP fusion gene. Shown are the N-terminal region cleaved by Kex2p protease after translation (hatched box), the cleavage site recognized by Kex2p protease (triangle), eight units of the repetitive sequence (shaded boxes), and the mRFP gene (black box). (B) The position of bud scars stained with CFW is identical to that of Pir1p. The Pir1p-mRFP fusion protein was expressed under the control of thePIR1promoter in JTS1. The images were taken with a BX50 fluorescence microscope and a MicroMAX cooled CCD camera.
FIG. 2. Pir1p localizes inside the chitin ring. The chitin rings of bud scars were stained with CFW (A) or FITC-WGA (B). The Pir1p-mRFP fusion protein was expressed under the control of thePIR1 promoter in JTS1. The images were taken using an IX71 fluorescence microscope with a confocal scanner and an EVM285SPD cooled CCD camera.
FIG. 3. Pir1p in diploid cells also localizes inside the bud scars. The chitin rings of bud scars were stained with CFW. The Pir1p-mRFP fusion protein was expressed under the control of thePIR1promoter in thePIR1disruptant (JTSD-1m). The images were taken using an IX71 fluorescence microscope with a confocal scanner and an EVM285SPD cooled CCD camera.
on September 8, 2020 by guest
http://ec.asm.org/
cells. Cell separation is facilitated by a partial digestion of the primary septum by a chitinase, and bud scars are the remains of the septum formed during these processes (see reference 3
for a review).CHS3encodes the putative catalytic subunit of
CSIII, andCHS3mutants do not exhibit a ring structure at bud
emergence (35). CHS2 encodes CSII, which is specific for
primary septum formation (35). Thechs2mutants also exhibit
an abnormal pattern of separation due to the lack of the primary septum, but the chitin ring at bud emergence is formed normally (34, 35). If Pir1p recognizes the primary septum or
the chitin ring, Pir1p-mRFP is expected to be invisible inchs2⌬
and chs3⌬ cells, respectively. Therefore, we investigated the
localization of Pir1p both inchs2⌬and chs3⌬cells. The ring
structure of Pir1p-mRFP was observed at the chitin rings in
chs2⌬cells (Fig. 4A). In contrast, although Pir1p-mRFP was
not detected as a ring structure, it was still observed as a cluster
at the budding site in chs3⌬ cells (Fig. 4B), suggesting that
Pir1p is transported to bud scars but cannot form a ring struc-ture because of the lack of a chitin ring. This result indicates that Pir1p is transported to the budding site and is located there even when the primary septum formation or the chitin ring is not normal.
Pir2p also localizes at bud scars.Next, we investigated the localization of Pir2p. Pir2p was fused with mRFP at its N terminus (mRFP-Pir2p) (Fig. 5A). The fusion site was placed downstream of the site cleaved by Kex2p because Pir2p fused with mRFP at its C terminus (Pir2p-mRFP) was not detected at the cell surface, probably due to degradation of the fusion protein (data not shown). The fusion protein mRFP-Pir2p was expressed on the cell surface, but some of it was concentrated into a unique circle in the cell wall, suggesting that Pir2p is also present in bud scars. As shown in Fig. 5B, the
ring structures were observed in cells expressing mRFP-Pir2p, confirming the localization of Pir2p at bud scars.
We also investigated the distribution of Pir3p and Pir4p in detail. Pir3p and Pir4p were fused with mRFP at the N and C terminus, respectively (Fig. 6A). As a result, both proteins were uniformly expressed on the cell surface irrespective of bud scars (Fig. 6B and 6C). The 3-D imaging of mRFP-Pir3p and Pir4p-mRFP revealed that these proteins are distributed on the cell surface almost uniformly and are not likely to be related to bud scars (data not shown).
Disruption of genes involved in bud site selection or cell polarity does not affect the localization of Pir1p.The budding site in haploid cells has been reported to be different from that
in diploid cells. Thea and␣ haploid cells bud with an axial
pattern, choosing new bud sites adjacent to the previous site of
bud emergence, whilea/␣diploid cells bud in a bipolar pattern,
choosing new bud sites at either end of the cell (8). The position where a new bud emerges on the cell surface is
de-termined by several factors. Mutations inBUD3,BUD4,AXL1,
orAXL2/BUD10cause a defect in the axial budding pattern in haploid cells, but they have no effect on the bipolar budding pattern of diploid cells (6, 7, 11, 12, 33). In contrast, mutations
inBNI1,SPA2,PEA2,BUD6,BUD7,BUD8, orBUD9in
dip-loid cells cause a defect in the bipolar budding pattern but have
FIG. 4. Pir1p is localized at the budding site both inchs2⌬cells (A) and inchs3⌬cells (B). The Pir1p-mRFP fusion protein was expressed under the control of thePIR1promoter inpir1⌬chs2⌬andpir1⌬chs3⌬ cells, respectively. The ring structure of Pir1p-mRFP was not observed in chs3⌬cells due to the lack of a chitin ring. The chitin rings of bud scars were stained with CFW. The images were taken with a BX50 fluorescence
microscope and a MicroMAX cooled CCD camera. region cleaved by Kex2p protease after translation (hatched box), theFIG. 5. (A) mRFP-PIR2fusion gene. Shown are the N-terminal cleavage site recognized by Kex2p protease (triangle), the 11 units of the repetitive sequence (shaded boxes), and the mRFP gene (black box). (B) Pir2p was expressed both on the cell surface (upper left panel) and at the bud scars (upper right panel). Both upper panels show the same cells expressing mRFP-Pir2p on different focal planes. The chitin rings of bud scars were stained with FITC-WGA. The mRFP-Pir2p fusion protein was expressed under the control of the PIR2promoter in JTS2. The images were taken with a BX50 fluores-cence microscope and a MicroMAX cooled CCD camera.
on September 8, 2020 by guest
http://ec.asm.org/
no effect on the axial budding pattern (2, 13, 43, 47). There-fore, it is possible that the disruption of these genes may affect the localization of Pir1p at bud scars both in haploid and
diploid cells. Consequently, we individually disrupted BNI1,
SPA2, PEA2, AXL2, BUD3, and BUD8 in strain JTS1-1m,
which produces mRFP-tagged Pir1p. In addition, we disrupted
several genes related to cell polarity:RGA1, a gene encoding a
GTPase-activating protein for the polarity-establishment
pro-tein Cdc42p (39);BEM1, a gene involved in normal bud
emer-gence and morphogenesis (23); andBEM2, a gene encoding a
Rho GTPase-activating protein involved in the control of the cytoskeletal organization and cellular morphogenesis (25, 45). However, disruption of these genes had no effect on the
local-ization of Pir1p (data not shown). Thebud10⌬cells, for
exam-ple, exhibited a bipolar pattern of bud site selection even in haploid cells, but Pir1p was observed at the incorrectly posi-tioned bud scars. These results suggested that Pir1p localizes to bud scars independent of cell polarity and bud site selection.
Next, we disrupted two genes that reportedly associate with
Pir1p: VMA6, which encodes vacuolar ATPase V0 domain
subunit d (14); andAPN1, which encodes a key enzyme in the
base excision repair pathway in the nucleus and mitochondria (44). However, neither gene disruption had any effect on the localization of Pir1p (data not shown).
Repetitive sequence of Pir1p is essential for its binding to cell wall.The repetitive sequence of Pir1p consists of eight tan-dem units of 18 to 19 amino acids. To investigate the relationship between this repetitive sequence and the localization to bud scars,
we removed these units stepwise from the N terminus. We cre-ated six types of Pir1p derivatives: Pir1p-A, Pir1p-B, Pir1p-C, Pir1p-D, Pir1p-E, and Pir1p-F. These proteins contain five, four, three, two, one, and no repetitive sequence units, respectively (Fig. 7A), and each was fused with mRFP at its C terminus. As shown in Fig. 7B, Pir1p-A, Pir1p-B, Pir1p-C, Pir1p-D, and Pir1p-E were localized at bud scars, but Pir1p-F was not detected anywhere on the cell surface.
Pir1p-F, which does not contain any repetitive sequence units, may not bind to the cell wall but may instead be released into the culture medium. Thus, we measured the amount of Pir1p-mRFP or its truncated derivatives that were secreted in the medium. Western blotting revealed that the amount of secreted Pir1p gradually increased as the number of repetitive units decreased (Fig. 8A). These fusion proteins appeared as smears with molecular weights higher than their predicted protein sizes, implying that they are highly glycosylated. Pir1p-E and Pir1p-F were smaller than the other Pir1p-mRFP fusion proteins due to the lack of one N-linked sugar chain (Fig. 7A). These results indicated that the repetitive sequence plays some role in the binding of Pir1p to the cell wall. It is also likely that the Pir1p derivatives containing many repetitive sequence units can bind strongly to the cell wall, whereas the
FIG. 6. (A) mRFP-PIR3 and PIR4-mRFP fusion genes. Shown are the N-terminal region cleaved by Kex2p protease after translation (hatched box), the cleavage site recognized by Kex2p protease (trian-gle), the eight units (Pir3p) or one unit (Pir4p) of the repetitive se-quence (shaded boxes), and the mRFP gene (black box). (B) Local-ization of mRFP-Pir3p. The mRFP-Pir3p fusion protein was expressed under the control of thePIR3promoter in JTS3. (C) Localization of Pir4p-mRFP. The Pir4p-mRFP fusion protein was expressed under the control ofPIR4promoter in JTS4. The images shown in panels B and C were taken with a BX50 fluorescence microscope and a MicroMAX cooled CCD camera.
FIG. 7. (A) Stepwise deletion of the repetitive sequence of Pir1p. Shown are the N-terminal region cleaved by Kex2p protease (hatched box), the cleavage site recognized by Kex2p protease (triangle), the repetitive units (shaded boxes), and the mRFP gene (black box). The circles indicate N glycosylation sites. (B) Localization of deleted ver-sions of Pir1p-mRFP (left panels) and CFW staining (right panels). The images were taken with a BX50 fluorescence microscope and a MicroMAX cooled CCD camera.
on September 8, 2020 by guest
http://ec.asm.org/
Pir1p derivative containing one repetitive unit has little ability to bind to the cell wall.
Next, we measured the amount of Pir1p on the cell wall. First, we tried to prepare Pir1p-mRFP from the isolated cell wall fractions by alkaline treatment as reported previously (1). However, almost all Pir1p-mRFP fusion proteins were de-graded, probably due to the vigorous breaking of yeast cells with glass beads during the preparation of the cell wall frac-tion. To attempt to avoid the degradation of Pir1p-mRFP, we prepared it from intact whole cells. Yeast cells expressing Pir1p-mRFP or its truncated derivatives Pir1p-A to Pir1p-F were cultivated, collected, washed, and then resuspended in a mild alkali solution. Western blotting confirmed the isolation of Pir1p-mRFP (Fig. 8B). In addition, Pir1p-A, Pir1p-B, Pir1p-C, Pir1p-D, and Pir1p-E were detected, but Pir1p-F was not. Although we were unable to quantify the results shown in Fig. 8A, the amounts of Pir1p-A to Pir1p-E released from the cell wall were obviously higher than the amount of Pir1p-mRFP (Fig. 8B). Because each of these proteins was expressed under its own promoter and the total amount of expressed protein appeared to be identical, this suggested that the deriv-atives were easily released from the cell wall due to their shorter repetitive sequence. Furthermore, the inability to de-tect Pir1p-F indicates that little if any of it localizes to the cell wall, which is consistent with our microscopic observations (Fig. 7B).
The C-terminal region of Pir1p is required for its recruit-ment to bud scars.To investigate whether the repetitive se-quence of Pir1p is involved in its recruitment to bud scars, we replaced the repetitive sequence of Pir4p, which distributes uniformly on the cell wall (Fig. 6C), with that of Pir1p, creating the chimeric protein Pir4/Pir1 (Fig. 9A). Pir4p has only one repetitive unit, and it was reported that this sequence is nec-essary for its binding to the cell wall (5). The chimeric protein with the repetitive sequence of Pir4p followed by the C-termi-nal region of Pir1p was fused with mRFP at its C terminus (Fig. 9A). As shown in Fig. 9B, this chimeric protein localized
to bud scars, indicating that the region downstream from the repetitive sequence of Pir1p, not the repetitive sequence itself, is important for its recruitment to bud scars.
DISCUSSION
Advantage of 3-D microscopic observation of yeast cells.In the present studies, 3-D superfine fluorescence microscopy allowed the detection of Pir1p in more detail and the collection of more information than two-dimensional fluorescence mi-croscopy. Because all of the Pir-mRFP fusion proteins were expressed under the control of their own promoters and all
PIR-mRFP fusion genes were integrated into the chromosome,
the fluorescence derived from Pir-mRFP fusion proteins could be used as an indication of the amount of native protein in the cells. Because of this ability of superfine 3-D microscopic sys-tems, they will become increasingly useful for detecting the precise cellular localization of proteins. For example, 3-D flu-orescence microscopy allowed us to determine that Pir1p is not always located inside the chitin ring of bud scars (see the supplemental material at http://unit.aist.go.jp/rcg/rcg-gb/pir1 .html). This result suggests that Pir1p is localized and functions in the bud scars after cell separation. This is important infor-mation for elucidating the function of Pir1p. Furthermore, without 3-D fluorescence microscopic systems, it is quite dif-ficult to detect all bud scars in a whole cell, and it is unlikely that the same results would have been obtained using conven-tional two-dimensional confocal microscopy. Therefore, it may be interesting to investigate the cellular localization of other proteins with 3-D fluorescence microscopic systems even if their localization has already been reported. However, our current 3-D fluorescence microscopic system still has some
FIG. 8. (A) Western blotting to detect the secreted Pir1p-mRFP. Lanes: W, wild type withoutPIR1-mRFP; Pir1p-mRFP, JTS1-1m; A, JTS1-A; B, JTS1-B; C, JTS1-C; D, JTS1-D; E, JTS1-E; F, JTS1-F. (B) Western blotting to detect the cell wall-localized Pir1p-mRFP released from the strains shown in Fig. 7 by a mild alkali treatment. Lanes: W, wild type withoutPIR1-mRFP; Pir1p-mRFP, JTS1-1m; A, JTS1-A; B, JTS1-B; C, JTS1-C; D, JTS1-D; E, JTS1-E; F, JTS1-F.
FIG. 9. (A) A chimeric Pir1p containing the repetitive sequence of Pir4p instead of the Pir1p repetitive sequence (Pir4/Pir1 chimeric protein) was produced under the expression of thePIR1promoter. Shown are the N-terminal region of Pir1p cleaved by Kex2p protease (hatched box), the cleavage sites recognized by Kex2p protease (tri-angle), the Pir4p repetitive sequence (shaded box), and the mRFP gene (black box). (B) The Pir4/Pir1 chimeric protein fused with mRFP was localized to bud scars (middle panel). The chitin ring was stained with CFW (left panel), and a phase-contrast view of cells is shown (right panel). The images were taken with a BX50 fluorescence mi-croscope and a MicroMAX cooled CCD camera.
on September 8, 2020 by guest
http://ec.asm.org/
limitations. For example, it took more than 1 min to collect the 100 FITC and mRFP confocal images for the view shown in the supplemental material. Therefore, it will be difficult to identify the cellular localization of quickly moving proteins using cur-rent 3-D systems. Further improvement of our prototype 3-D fluorescence microscopic system should provide more precise information not only for Pir proteins but also for other proteins of interest.
Function of Pir1p.Although thePIRgenes were identified by Toh-e and coworkers in 1993 (42), the function of the Pir proteins and the role of their repetitive sequences have not yet been determined. Here, we reported that Pir1p is localized inside the chitin ring of bud scars. For more than 30 years, the bud scar has been known as a ring-like structure made almost entirely of chitin, but other components of the chitin ring have not been investigated. Pir1p is the first protein shown to be specifically expressed inside bud scars. Pir2p is also located at bud scars, but it exists in other regions of the cell wall as well. Pir3p and Pir4p are uniformly expressed in the entire cell wall and are not localized at bud scars. Moukadiri et al. reported that Pir4p is distributed predominantly on the surface of grow-ing daughter cells (27). They observed Pir4p by usgrow-ing an indi-rect immunofluorescence method with a polyclonal antibody against Pir4p. The discrepancy in the localization of Pir4p may be due to the differences in the method of visualization.
The difference in cellular localization among Pir family pro-teins may reflect a difference in their functional positions. Because Pir1p is not localized at the bud neck during the budding process but is present at bud scars after the comple-tion of cell separacomple-tion, it is conceivable that Pir1p is required for reinforcement of the weakened cell wall following cell budding. If this is the case, Pir3p and Pir4p may have some function that compensates or protects the entire cell wall from the various forms of environmental stress. Pir2p may reinforce the cell wall both at bud scars and in other regions. In fact, the
yeast cell compensates for the decreased level of-1,6-glucan
in its cell wall by upregulating the transcription of Pir (19, 37). In addition, it was reported that a lack of some Pir proteins renders cells sensitive to heat (42) and tobacco osmotin (46). It
was also reported that cells with disruptedPIRgenes showed
growth defects in the presence of CFW or Congo red (30). These results support our hypothesis that Pir proteins maintain
the integrity of the cell wall. Because PIR1 and PIR2 are
reported to be highly expressed during G1 phase (38), Pir1p
and Pir2p may be incorporated in bud scars to reinforce the budding site after the completion of the budding process.
Because Pir1p is the first protein shown to be localized to bud scars, it may have functions other than cell wall reinforce-ment. For example, it is possible that Pir1p acts as a marker to prevent new budding from a site where budding was just com-pleted. It is also possible that Pir1p expands and/or breaks the chitin ring in aged cells, a process that occurs as the cell grows (32). Further analysis is required to understand the precise role of Pir proteins.
Role of the repetitive sequence of Pir proteins.The repeti-tive sequence of Pir proteins was first investigated by Castillo et al. using Pir4p, which has only one repetitive unit (5). Evi-dence was obtained that the repetitive sequence of Pir4p is necessary for its anchoring to the cell wall. Because Pir1p was found to localize to bud scars, we investigated the functional relationship between the repetitive sequence and the localiza-tion of Pir1p. The finding that a chimeric protein consisting of the repetitive sequence of Pir4p and the C-terminal region of Pir1p is present at bud scars provided us with an important clue. This result clearly demonstrates that the repetitive se-quence of Pir1p is required for the binding to the cell wall and that the C-terminal region is necessary for recruitment to spe-cific regions of the cell wall. Considering the previous results (5), we expect that the repetitive sequence of Pir proteins anchors the proteins to the cell wall. However, it is still unclear how Pir proteins are cross-linked to the cell wall through their repetitive sequences. Their release from cell walls by a mild alkaline treatment suggests that they may be cross-linked through their O-glycans. Thus, it is likely that Pir1p is cross-linked to the cell wall through O-glycans attached to the re-petitive sequences.
Mechanism of Pir protein localization to bud scars. How and when Pir1p is translocated to bud scars is unknown. Be-cause Pir1p is not detected around the bud neck and beBe-cause the bud scars do not always contain Pir1p, we expect that Pir1p translocates to bud scars after the completion of cell separa-tion. The precise localization of Pir1p in bud scars is also unclear. To distinguish whether Pir1p recognizes the primary septum or the chitin ring, we investigated the localization of
Pir1p inchs2⌬andchs3⌬cells because the primary septum and
the chitin ring are not formed normally inchs2⌬ and chs3⌬
FIG. 10. An alignment of the C-terminal regions of Pir1p to Pir4p. The sequences highly conserved in Pir1p and Pir2p but not in Pir3p and Pir4p are indicated by asterisks. The numbers correspond to the amino acid residues of the Kex2p-processed form of each Pir protein.
on September 8, 2020 by guest
http://ec.asm.org/
cells, respectively. We first expected that if Pir1p recognizes the primary septum or the chitin ring, Pir1p is not detected at
bud scars in chs2⌬ and chs3⌬ cells, respectively. However,
Pir1p is translocated to bud scars both inchs2⌬andchs3⌬cells
(Fig. 4). The different appearance of Pir1p inchs2⌬andchs3⌬
cells may be due to the different morphology of the bud scars in these mutants. This finding indicates that Pir1p is translo-cated to the budding site even when septum formation or the chitin ring is not normal. Further investigations, for example by immunoelectron microscopy, are necessary to determine the precise location of Pir1p. This information and determination of the period when Pir1p is localized to bud scars will lead to a better understanding of Pir1p function.
We disrupted several genes involved in bud site selection and cell polarity. As expected, due to the disruption of these genes, budding patterns and cell polarity became abnormal. However, Pir1p still localized at incorrectly positioned bud scars. These results indicate that Pir1p is transported and localized to bud scars independent of the mechanisms of cell polarity and bud site selection. We expect that there is an as-yet-unidentified mecha-nism for transporting proteins such as Pir1p to bud scars. The truncation of Pir1p indicates that the region responsible for its recruitment to bud scars lies in the C terminus. Combined with the finding that Pir2p is also transported to bud scars, this result indicates that Pir1p and Pir2p may have the same or similar signal sequences in their C termini for transport to bud scars. An align-ment of the amino acid sequences downstream from the repeti-tive sequences of Pir1p to Pir4p is shown in Fig. 10. Although the C-terminal regions of the four Pir proteins are similar, Pir1p and Pir2p have highly conserved sequences that are not conserved in Pir3p and Pir4p. It is likely that the amino acids around this sequence act as a bud scar localization signal, although a more detailed identification of the targeting sequence will be necessary.
ACKNOWLEDGMENTS
We are grateful to Roger Tsien (Howard Hughes Medical Institute Laboratories, University of California at San Diego) for providing the mRFP gene and to K. Nakayama, Y. Chiba, X. Gao, and H. Abe for helpful discussions.
This work was partly supported by the Japan Society for the Pro-motion of Science (JSPS) and a grant-in-aid for the DB project by the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
REFERENCES
1.Abe, H., Y. Shimma, and Y. Jigami.2003. In vitro oligosaccharide synthesis using intact yeast cells that display glycosyltransferases at the cell surface through cell wall-anchored protein Pir. Glycobiology13:87–95.
2.Amberg, D. C., J. E. Zahner, J. W. Mulholland, J. R. Pringle, and D. Botstein.1997. Aip3p/Bud6p, a yeast actinteracting protein that is in-volved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell8:729–753.
3.Cabib, E., D.-H. Roh, M. Schmidt, L. B. Crotti, and A. Varma.2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem.276:19679–19682.
4.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien.2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA99:7877–7882.
5.Castillo, L., A. Martinez, A. Garcera, V. Elorza, E. Valentin, and R. Sentandreu.2003. Functional analysis of the cysteine residues and the re-petitive sequence ofSaccharomyces cerevisiaePir4/Cis3: the repetitive se-quence is needed for binding to the cell wall-1,3-glucan. Yeast20:973–983. 6.Chant, J., and I. Herskowitz.1991. Genetic control of bud-site selection in yeast by a set of gene products that comprise a morphogenetic pathway. Cell
65:1203–1212.
7.Chant, J., K. Corrado, J. R. Pringle, and I. Herskowitz.1991. YeastBUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell65:1213–1224.
8.Chant, J., and J. R. Pringle.1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol.129:751–765.
9.De Groot, P. W. J., A. F. Ram, and F. M. Klis.2005. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol.42:657– 675.
10.Doolin, M. T., A. L. Johnson, L. H. Johnston, and G. Butler.2001. Over-lapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol.40:422–432.
11.Fujita, A., C. Oka, Y. Arikawa, T. Katagai, A. Tonouchi, S. Kuhara, and Y. Misumi.1994. A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature372:567–570. 12.Halme, A., M. Michelitch, E. L. Mitchell, and J. Chant.1996. Bud10p directs
axial cell polarization in budding yeast and resembles a transmembrane receptor. Curr. Biol.6:570–579.
13.Harkins, H. A., N. Page, L. R. Schenkman, C. De Virgilio, S. Shaw, H. Bussey, and J. R. Pringle.2001. Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol. Biol. Cell12:2497–2518. 14.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki.2001. A
comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA98:4569–4574.
15.Jaafar, L., I. Moukadiri, and J. Zueco.2003. Characterization of a disul-phide-bound Pir-cell wall protein (Pir-CWP) ofYarrowia lipolytica. Yeast
20:417–426.
16.Kandasamy, R., G. Vediyappan, and L. Chaffin.2000. Evidence for the presence of Pir-like proteins inCandida albicans. FEMS Microbiol. Lett.
186:239–243.
17.Kapteyn, J. C., R. C. Montijn, E. De Vink, J. La Cruz, A. Llobell, J. E. Douwes, H. Shimoi, P. N. Lipke, and F. M. Klis.1996. Retention of Sac-charomyces cerevisiae cell wall proteins through a phosphodiester-linked
-1,3-/-1,6-glucan heteropolymer. Glycobiology6:337–345.
18.Kapteyn, J. C., H. Van Den Ende, and F. M. Klis.1999. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta1426:373–383.
19.Kapteyn, J. C., P. Van Egmond, E. Sievi, H. Van Den Ende, M. Makarow, and F. M. Klis.1999. The contribution of theO-glycosylated protein Pir2/ Hsp150 to the construction of the yeast cell wall in wild-type cells and
-1,6-glucan deficient mutants. Mol. Microbiol.31:1835–1844.
20.Klebe, R. J., J. V. Harriss, Z. D. Sharp, and M. G. Douglas.1983. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene25:333–341.
21.Kollar, R., B. B. Reinhold, E. Petrakova, H. J. C. Yeh, G. Ashwell, J. Drgonova, J. C. Kapteyn, F. M. Klis, and E. Cabib.1997. Architecture of the yeast cell wall.-(136)-glucan interconnects mannoprotein,-(13 3)-glu-can, and chitin. J. Biol. Chem.272:17762–17775.
22.Lagorce, A., N. C. Hauser, D. Labourdette, C. Rodriguez, H. Martin-Yken, J. Arroyo, J. Hoheisel, and J. Francois.2003. Genome-wide analysis of the response to cell wall mutations in the yeastSaccharomyces cerevisiae. J. Biol. Chem.278:20345–20357.
23.Leeuw, T., A. Fourest-Lieuvin, C. Wu, J. Chenevert, K. Clark, M. Whiteway, D. Y. Thomas, and E. Leberer.1995. Pheromone response in yeast: associ-ation of Bem1p with proteins of the MAP kinase cascade and actin. Science
270:1210–1213.
24.Longtine, M. S., A. Mckenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle.1998. Additional modules for versatile and econominal PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast14:953–961.
25.Marquitz, A. R., J. C. Harrison, I. Bose, T. R. Zyla, J. N. McMillan, and D. J. Lew.2002. The Rho-GAP Bem2p plays a GAP-independent role in the morphogenesis checkpoint. EMBO J.21:4012–4025.
26.Martinez, A. I., L. Castillo, A. Garcera, M. V. Elorza, E. V. Valentin, and R. Sentandreu.2004. Role of Pir1 in the construction of theCandida albicans cell wall. Microbiology150:3151–3161.
27.Moukadiri, I., L. Jaafar, and J. Zueco.1999. Identification of two manno-proteins released from cell walls of aSaccharomyces cerevisiae mnn1 mnn9 double mutant by reducing agents. J. Bacteriol.181:4741–4745.
28.Moukadiri, I., and J. Zueco.2001. Evidence for the attachment of Hsp150/ Pir2 to the cell wall ofSaccharomyces cerevisiaethrough disulfide bridges. FEMS Yeast Res.1:241–245.
29.Mrsa, V., T. Seidl, M. Gentzsch, and W. Tanner.1997. Specific labeling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins ofSaccharomyces cerevisiae. Yeast13:1145–1154. 30.Mrsa, V., and W. Tanner.1999. Role of NaOH-extractable cell wall proteins Ccw5p, Ccw6p, Ccw7p and Ccw8p (members of Pir protein family) in sta-bility of theSaccharomyces cerevisiaecell wall. Yeast15:813–820. 31.Orlean, P., H. Ammer, M. Watzele, and W. Tanner.1986. Synthesis of an
O-glycosylated cell surface protein induced in yeast by␣-factor. Proc. Natl. Acad. Sci. USA83:6263–6266.
32.Powell, C. D., D. E. Quain, and K. A. Smart.2003. Chitin scar breaks in aged Saccharomyces cerevisiae. Microbiology149:3129–3137.
33.Roemer, T., K. Madden, J. T. Chang, and M. Snyder.1996. Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev.10:777–793.
on September 8, 2020 by guest
http://ec.asm.org/
34.Schmidt, M., B. Bowers, A. Varma, D.-H. Roh, and E. Cabib.2002. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci.115:293– 302.
35.Shaw, J. A., P. C. Mol, B. Bowers, S. J. Silverman, M. H. Valdivieso, A. Duran, and E. Cabib.1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol.114:111–123.
36.Sherman, F.1991. Getting started with yeast. Methods Enzymol.194:3–21. 37.Smits, G. J., J. C. Kapteyn, H. Van Den Ende, and F. M. Klis.1999. Cell wall
dynamics in yeast. Curr. Opin. Microbiol.2:348–352.
38.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeastSaccharomyces cer-evisiaeby microarray hybridization. Mol. Biol. Cell9:3273–3297. 39.Stevenson, B. J., B. Ferguson, C. De Virgilio, E. Bi, J. R. Pringle, G.
Am-merer, and G. F. Sprague, Jr.1995. Mutation ofRGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharo-myces cerevisiae. Genes Dev.9:2949–2963.
40.Teparic, R., I. Stuparevic, and V. Mrsa.2004. Increased mortality of Sac-charomyces cerevisiaecell wall protein mutants. Microbiology150:3145–3150.
41.Thomas, B. J., and R. Rothstein.1989. Elevated recombination rates in transcriptionally active DNA. Cell56:619–630.
42.Toh-e, A., S. Yasunaga, H. Nisogi, K. Tanaka, T. Oguchi, and Y. Matsui.
1993. Three yeast genes,PIR1,PIR2andPIR3, containing internal tandem repeats, are related to each other, andPIR1 andPIR2 are required for tolerance to heat shock. Yeast9:481–494.
43.Valtz, N., and I. Herskowitz.1996. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J. Cell Biol.135:725–739.
44.Vongsamphanh, R., P. K. Fortier, and D. Ramotar.2001. Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol. Cell. Biol.21:1647–1655.
45.Wang, T., and A. Bretscher.1995. The rho-GAP encoded byBEM2regulates cytoskeletal structure in budding yeast. Mol. Biol. Cell6:1011–1024. 46.Yun, D. J., Y. Zhao, J. M. Pardo, M. L. Narasimhan, B. Damsz, H. Lee, L. R.
Abad, M. P. D’Urzo, P. M. Hasegawa, and R. A. Bressan. 1997. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc. Natl. Acad. Sci. USA94:7082–7087.
47.Zahner, J. E., H. A. Harkins, and J. R. Pringle.1996. Genetic analysis of the bipolar pattern of bud site selection in the yeastSaccharomyces cerevisiae. Mol. Cell. Biol16:1857–1870.