CopyrightC1976 AmericanSociety for Microbiology PrintedinU.SA.
Construction
of the
Genetic
Map
of the
Polyoma
Genome
LOIS K. MILLER* AND MIKE FRIED
Imperial CancerResearchFund,Lincoln's InnFields,London WC2A3PX, England
Received for publication26January 1976
Sevenearly mutants, three late mutants, andoneplaquemorphology mutant
of polyoma have been mapped by marker rescue using wild-type restriction endonuclease fragments. The early mutants map between 1.0 and 26.4 units from the Eco RI site, a region previously shown to correspond to the 3'-OH
terminal halfof"early"RNA(Kamenetal., 1974). Thelatemutantsaswellas
the plaque morphologymutantmapbetween26.6and 45.4mapunits,aregion
previously shown to correspond to the 3'-OH terminal half of "late" RNA
(Kamen etal., 1974). Analysis of thegenotype ofrescuedvirus demonstrated
that the modificationofthemutantDNAduringmarkerrescuewaslimitedto
the region of the genome covered by the wild-type restriction endonuclease
fragment tested.
Temperature-sensitive mutants of the onco-genic papovavirus, polyoma,havebeenisolated and characterized (2-4, 7, 8). These mutants generally fall intotwoclasses: (i)earlymutants that areblockedinviral DNAsynthesis (6, 7) atthe restrictive temperature, and (ii) late mu-tants that do not form infectious particles al-though viral DNA issynthesizedatthe restric-tive temperature (2-4). Genetic complementa-tionanalysis ofthe mutants has provided some further subdivisions in this classification scheme (2-4), but adetailed knowledge ofthe genetic organization ofpolyoma virus has re-mained elusive due to low recombination fre-quencies (14) and lack of sufficiently distinct
conditional-lethal mutation types for genetic mappingby three-factor genetic crosses.
The genetic mapping technique of marker rescue is anovel one since itinvolves the bio-chemical manipulation of the viral DNA. The
technique relies on the ability of a fragment of
wild-type DNA, which has no infectivity by itself, to produce wild-type virus when an-nealedto afull-lengthmutant DNAand intro-duced to a cell. This will be referred to as a "frescue"p event, but it is important to note that the original mutant DNA is converted to a wild-type DNA by cellular mechanisms.
The rescue ofwild-type virusfrom
transfec-tions with heteroduplex DNA forned by an-nealing wild-type DNA fragments to condi-tional-lethal mutant viral DNA was first ob-served in kX174 studies (13, 28). The marker
rescue technique has been shown to be an
effective method for mapping mutants of sim-ianvirus40(SV40) (16-18). The marker rescue procedure relies on theability to produce
spe-cificfragments of viral DNA by cleavage with
restriction endonucleases, to purify the
frag-ments by gelelectrophoresis, and to know the
physical orderof thefragmentswithin the ge-nome.
Anumberofrestriction endonucleases cleave polyoma DNA into discrete fragments, the physical order of which has been determined (10, 11;B.E. Griffin, unpublished data).Inthis
report, individual restriction endonuclease
fragments(RENFs)were testedfor theirability
to rescue avarietyofearlyandlatetsmutants.
A
genetic.
map of these mutations wascon-structedfrom the results. Inaddition,wetested
the ability of fragments to convert a minute-plaque mutantofpolyoma tothe large-plaque
type,andthe results demonstratethatthis
mu-tation is located in the "late" region of the genome.
MATERIALSAND METHODS
Viruses. All virus stockswerepreparedfrom
sin-gle plaquesand were grown at low multiplicities as
previouslydescribed (9). The early mutant tsa and the latemutanttsCwerederived from the Pasadena large-plaque strain byM.Fried (7; M.Fried, unpub-lisheddata). The CR variant was derived from the Pasadena large-plaque strain (Fried, unpublished
data).Theearly mutants ts25, ts616, ts48,ts609,and
ts697 and the late mutants tslO and ts1260 were providedbyW. Eckhart (4 and references therein). The ts697 mutant was derived from the P16 small-plaque strain, and the others were derived from the Pasadena large-plaque strain. The minute-plaque mutant, 208,wasderived from the Pasadena large-plaquestrainandprovided byM.Vogt.
Cells and plaqueassay..Monolayersofsecondary wholemouse embryo cellsweregrowninDulbecco modified Eagle medium supplemented with 5% fetal 824
on November 10, 2019 by guest
http://jvi.asm.org/
POLYOMA GENETIC MAP 825
calfserum for viral DNA preparation or 3% horse serum for virus stock production or plaque assay. Plaque assays were performed as previously de-scribed (8).
Preparation of polyoma DNA. All viral DNAs
were prepared by Hirt extraction (12) of infected cells followed by CsCl-ethidium bromide equilib-rium density centrifugation (23). The supercoiled form I (FI) polyoma DNA was further purified by neutral sucrose gradient centrifugation. Random, singly nicked form II (FII) DNA was prepared by X
irradiationof FI tonick approximately 30% of the
molecules. The FII singly nicked DNAs were puri-fiedfrom the remaining FI DNA byCsCl-ethidium bromideequilibrium densitycentrifugation.
PreparationofRENFsofDNA. RENFs were
pre-pared from 3H-labeled FI DNA except the HpaII
fragments, which were prepared from32P-labeledFI DNA. The 3H-labeled fragments were purified by electrophoresis through 1.4% agarose tube gels, 20 cm inlength. The DNA fragments were visualized byethidiumbromidestaining(21, 24), cut individ-ually from the gels, andremoved from the agarose
by electrophoresis. The fragments werejudgedby
gel electrophoresis tobe more than 90% pure. The
32P-labeledHpaIIfragments (a gift of B. E. Griffin) werepurified byelectrophoresison3.3%
polyacryl-amide plus0.5%agaroseslabgels, locatedby
auto-radiography, slicedfrom the gels, andremovedfrom
thegelslicesbyelectrophoresis. Eachfragmentwas
furtherpurified by neutralsucrosegradient
centrif-ugationandcheckedfor puritybygel electrophore-sis.
Restrictionendonucleasesandincubation
condi-tions. Therestriction endonuclease HindIII was
pre-pared by proceduressimilar to thosedescribed
pre-viously (27). The HhaI enzyme waspurified bythe
unpublished procedure ofR. RobertsandP. Myers
(personal communication). TheHpaII and Eco RI
enzymes werekindlyprovided byB. GriffinandR. Kamen. The buffers used for enzyme digestions were: HhaI andHpaII, 10 mM Tris-hydrochloride (pH 7.5)-10 mM MgCl2-1 mM dithiothreitol;
HindIII, sameasHhaIandHpaTI + 50 mMNaCl;
Eco RI, 10mMTris-hydrochloride (pH 7.5)-10 mM
Mg2Cl-100mMNaCl.Incubationswere at 37 Cfor a
sufficienttimeandwithsufficientenzymefor
com-plete digestion. Thecleavage sitesof these
endonu-cleasesareshowninFig. 2(adaptedfrom reference
11).
Transfection of cells with polyomaDNA.
Near-confluentmonolayersofsecondarywhole mouse em-bryocells in 35-mmpetridisheswerewashedonce with 2 ml of Dulbeccophosphate-bufferedsaline
mi-nusMg2+andCa2+ (PBSA).A0.4-mlaliquotof1mg
ofDEAE-dextran per mlof PBSA wasdistributed
overthecellmonolayerandincubated at32C for20
min. The excessDEAE-dextran wasremoved,anda 0.3-ml solution ofpolyomaDNAinPBSAwas dis-tributed over the cell monolayer. The cells were
incubated forapproximately 30minat 32C, after
which 1.5 ml of Dulbecco modifiedEagle medium
supplemented with 3% horse serum and
receptor-destroying enzyme (8) was added. The cells were
incubatedat 32 C forapproximately6days.
Freeze-thawingfollowedby 0.5 min ofgentlesonicationwas usedtobreak open theinfectedcells.
FormationofheteroduplexDNAs. Heteroduplex
DNAs were formed by combining random, singly nicked, FII mutantpolyoma DNA, RENFs produced by cleavage of FI mutant DNA, and a single, puri-fiedRENF of wild-type polyoma DNA. In the first series ofexperiments(usingHpaHI fragments), the molar ratios of the various DNA types were
1:10:approximately250. In otherexperiments(using
HindIII, HhaI, and HhaI + Eco RIfragments),the molar ratios of the DNAs were 1:4:40 (mutant FII DNA:RENFs containing sequences of the entire mutantgenome:purifiedwild-typeRENF). To a
40-IlI
volumecontaining0.0125 ,ug of mutant FII DNA and the appropriate concentrations ofmutant and wild-typefragments dissolvedin 10 mM Tris-hydro-chloride (pH 7.5) and 1 mM EDTA, 10 ,ul of2 M NaOH was added. Afterstanding at room tempera-ture for 20min, thesolution was neutralized by the addition of 20 ,ul of a buffered HCIsolution (1 vol-ume of 2 MHCI, 1 volume of 1 M sodiumphosphate, pH 7.0). The neutralized solution was incubated at 68C for 30 min. Under these hybridization condi-tions, atleast 80% of thefull-lengthDNAmolecules are inthedouble-strandedform;approximatelyfour outof fivefull-length double-stranded DNA mole-cules contain onefragmented strand.After hybridi-zation, 0.1ml ofdistilledwaterand 0.45 ml of PBSA wereadded, and the solution was frozen until usedfortransfectionofcells. The DNAhybridizedinthis
mannershowedaspecificinfectivity of 2 x 105 PFU
per jig offull-length FU DNA by direct
DEAE-dextrantransfectionassayat 32 C.UntreatedFIor FII DNAhas aspecificinfectivity ofapproximately1 x 106 PFU per ugoTfDNA inDEAE-dextran trans-fection assays at 32 C.
RESULTS
Experimental design. Theexperimental de-sign forgenetic mapping ofpolyoma mutants
bymarkerrescueisschematicallypresented in Fig. 1.Heteroduplexeswereformedby combin-ing,denaturing,and reannealingmutant(ts or minute plaque) FII DNA, a single purified wild-type RENF, and the full complement of
mutantRENFsproduced byrestriction
endonu-clease cleavage of mutant FI DNA. The mu-tantRENFswereaddedfortworeasons: (i) to preventthereannealing offull-length mutant DNA strands of the added mutant FII DNA, and (ii) to minimize the enhancement of
marker rescue by nonspecific fragments that
Lai and Nathansobserved (16). Theresults of
Lai and Nathans demonstrated that a partial
heteroduplex composed ofafull-lengthmutant DNA and one wild-type restriction fragment
wasless effectiveinmarkerrescuethan a par-tialheteroduplex withmorethanonewild-type
restrictionfragment. Thisresultedin an
anom-alous enhancement ofinfectivityby
nonspecific
fragments, leading to some confusion as to VOL. 18, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
Mutant F1I Mutant Fragments One Wild-Type Fragment
Combine Denature
Reanneal
Heteroduplex
I Infect cells at 32 C Virus Stock
I Plaque Assay at 39C and 32 C
Ratio 39C/32C
FIG. 1. Experimentdesign for markerrescue.MutantFII DNA wascombined with thefullcomplementof mutant restrictionendonucleasefragments,produced byrestrictionnuclease digestion of FImutantDNA, andasingle, purifiedwild-type restriction nucleasefragment.The ratiosofthesethree DNA componentswere adjustedtopermit (i)thesinglewild-typefragmenttocompetesuccessfullywith a mutantfragment,and(ii) thefragments to compete successfully with the full-length mutantDNAduring the reannealing reaction. The threeDNAcomponentswere denatured and reannealed to allow the formationof heteroduplex circular DNA composed of one full-length mutant DNA and a complementary strandcomposed of onewild-typefragment andthe remainingmutant fragments. Whole mouse embryo cells were infected with this DNA at 32 C in the presence of receptor-destroying enzyme to inhibit secondary infections. The virus stock obtained from this infection (see Materials and Methods) was assayed for plaque-forming ability at 39 and 32 C. The wild-type restrictionendonuclease fragment resulting in the highest 39C/32C ratiocorresponds to the mutated region of the mutant DNA tested.
which wild-type fragment corresponded to the The molarratiosofthe threeDNA typesused
mutantregion. Weattemptedtominimizethis to form the heteroduplex were balanced in a
enhancement effect by startingwithafullhet- mannerthat (i)permittedthewild-typeRENF
eroduplexDNA. tocompetesuccessfullywiththecorresponding
826 MILLER AND
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.503.80.411.53.500.2]POLYOMAGENETIC MAP 827
mutantRENFsothatatleast9outof 10 hetero-duplexes contained the wild-type RENF, and (ii) permitted the RENFs to compete
success-fully with full-length DNA derived from the
mutantFII DNA toenhance formation of het-eroduplexes rather than homoduplexes.
Cells infected with the heteroduplex DNAs
wereincubatedat32 C inthepresenceof
recep-tor-destroying enzyme, which inhibits
reinfec-tion by the newly formed virus (8). This
pro-vided a stock ofmutant and wild-type virus,
which wasthenplaque assayedatthe
permis-sive and nonpermissive temperatures (32 and 39C). In the case ofthe minute-plaque virus,
208,theplaqueassayswereperformedat37 C,
andthenumber oflarge-plaqueviruses
appear-ing in the presence of minute plaques was
scored. Theratio of PFUassayedat39Ctothe PFU assayed at 32C (referred to asthe 39C/
32 Cratio)indicated which RENFcorresponded
to the ts region of the genome. Similarly, the
ratio oflarge plaqueto minuteplaqueat 37C indicated which RENF correspondedtothe
re-gion of the genome responsible for the plaque
morphology of themutanttested.
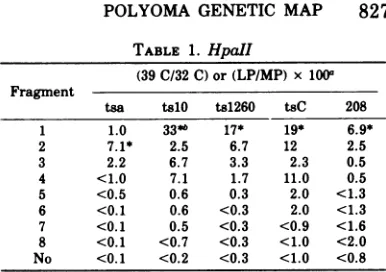
Marker rescue with HpaII fragments. In the first experiment for genetically mapping polyoma mutants, the eight HpaII RENFs of wild-type DNA were individually tested for
theirability to "rescue" theearly mutant tsa,
the late mutants ts1O, ts1260, tsC, and the plaque morphology mutant 208 (Table 1). For theearlymutant,tsa,themostactivefragment inmarkerrescue wasHpaII fragment2 (HpaII-2).Forthe latemutantstslO,ts1260and tsCas
wellastheplaque morphologymutant208,the most active fragment for marker rescue was
HpaII-1. Theseresults, however, are not
com-pletely satisfactory since other fragments
showed unusually high ratios (backgrounds).
The results with tsC areparticularly ambigu-ous. The fragments that showed high
back-groundswere usuallythosethatmigrated just
below themostpositive fragment.
For instance,inthetsacase,fragment3gave
aratioapproximatleyone-third the ratio forthe
most positive fragment (HpaII-2), whereas Hpall fragments 2 through4 gave high back-groundswhen tslO was tested. In thecases of tslOandtsC,theHpaII-4 fragmentgaveahigh
background. Although we have no complete
explanation for this observation, our results
using other RENFs demonstrate that the
mu-tantregionof thesetwomutantsis not located in the HpaH-4 region. The problem of high background wasprobably due to impuritiesof
fragment preparations. Since the Hpall
[image:4.503.260.453.57.193.2]frag-mentsweregreater than90%pureasjudged by
TABLE 1. HpaII
(39 C/32C)or(LP/MP)x 100"
Fragment
tsa talO ts1260 tsC 208
1 1.0 33*b 17* 19* 6.9*
2 7.1* 2.5 6.7 12 2.5
3 2.2 6.7 3.3 2.3 0.5
4 <1.0 7.1 1.7 11.0 0.5 5 <0.5 0.6 0.3 2.0 <1.3 6 <0. 1 0.6 <0.3 2.0 <1.3 7 <0.1 0.5 <0.3 <0.9 <1.6 8 <0.1 <0.7 <0.3 <1.0 <2.0 No <0.1 <0.2 <0.3 <1.0 <0.8
aValuesare averages of at least twoplates for at least
threedilutions over a rangeoftwoordersofmagnitudefor
both 39 C and 32 C. LP, Largeplaque;MP, minuteplaque. bAsterisk indicates themostpositive response.
gelelectrophoresis in the presence of ethidium
bromide andthe method of heteroduplex
forma-tionused wasdesigned to minimize the effect of nonspecific fragment enhancement of marker rescue, weconsidered that the effects of small contaminations in the fragment preparation were accentuated by the high ratios of frag-ments to mutant FII DNA used to form the heteroduplexes. In further experiments (Tables 2-5), we used molar ratios of mutant FII DNA to mutant RENFs to wild-type RENF of 1:4:40 informingheteroduplex DNAs rather than ra-tiosof1:10:250usedintheHpaIIexperiments of Table 1. This change generally improved the differences between the 39 C/32 C ratios of the various fragments (Tables 2-5). However, in
somecases westill observedhighbackgrounds,
andwesuspectthat thiswasduetothe nonspe-cific fragment enhancement effectreported by Lai and Nathans (16), even though we at-tempted to minimize this effect in our experi-mental design.
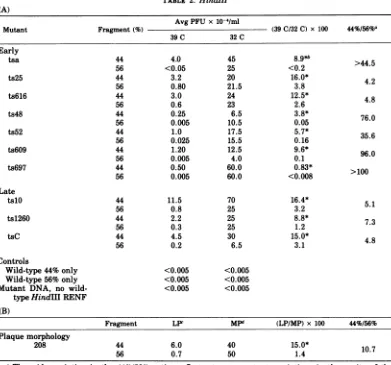
Marker rescue with HindIII fragments. The twofragmentsproducedbycleavage of polyoma DNA with HindIII (11) were tested for their
abilitytorescue avariety ofpolyomamutants. Every mutanttestedwaspreferentially rescued
by the smaller of the two fragments, which
contained approximately 44% of the polyoma
genome (Table2). These resultsareconsistent with the HpaII results (Table 1) since the HindIII-44% fragment contained large
seg-ments of both the HpaII-1 and HpaH-2
frag-ments, and demonstrate a clustering ofearly and late mutations into a restricted region
(44%) of the polyoma genome. These results have been further confirmed by testing
frag-mentsproduced by cleavagewith HhaI
restric-tionendonuclease (seebelow).
Marker rescue with HhaI + Eco RI frag-ments. Digestion of polyoma wild-type DNA with acombinationof HhaI and Eco RI
restric-VOL. 18, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
TABLE 2. HindlII (A)
Avg PFU x 10-4/ml
Mutant Fragment(%) (39C/32 C) x 100 44%/56%a
39C 32C
Early
tsa 44 4.0 45
8.9*>
56 <0.05 25 <0.2
ts25 44 3.2 20 16.0* 4.2
56 0.80 21.5 3.8
ts616 44 3.0 24 12.5* 4.8
56 0.6 23 2.6
ts48 44 0.25 6.5 3.8*
56 0.005 10.5 0.05 76.0
ts52 44 1.0 17.5 5.7* 3
56 0.025 15.5 0.16 5.6
ts609 44 1.20 12.5 9.6*
56 0.005 4.0 0.1 96.0
ts697 44 0.50 60.0 0.83*
56 0.005 60.0 <0.008 >100
Late
tslO 44 11.5 70 16.4* 5.1
56 0.8 25 3.2
ts1260 44 2.2 25 8.8* 7.3
56 0.3 25 1.2
tsC 44 4.5 30 15.0*
4.8
56 0.2 6.5 3.1
Controls
Wild-type 44%only <0.005 <0.005
Wild-type56%only <0.005 <0.005
Mutant DNA, no wild- <0.005 <0.005 typeHindIII RENF
(B)
Fragment LPr MP" (LP/MP) x 100 44%/56% Plaque morphology
208 44 6.0 40 15.0* 10.7
56 0.7 50 1.4
aThe wide variation in the 44%/56% ratios reflects, to some extent, variations in the purity of the
differentfragment preparations used to test each mutant. However, differencesinvirus strainsandinthe type orlocation of the mutation may also affect these ratios.
bAsteriskindicates most positive response.
cLP, Largeplaque.
d
MP,
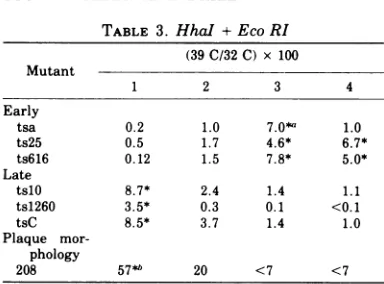
Minuteplaque.tionendonucleasesproducesfourfragments ap-proximately 46, 28, 14, and 12% (Fig. 2 and reference 10). These fragments are designated HhaI/EcoRI-1,-2,-3, and-4 inorder of
decreas-ing size.
The results of marker rescue using these HhaI+Eco RIfragmentsareshowninTable3. Of theearlymutants,tsa wasrescued by frag-ment 3, whereas ts25 and ts616 were rescued almostequallywellby fragments3and4.Since itisrelatively difficult to purify these two small fragments, another approach was used to
dem-onstrate that the sequences in fragment 4
rather than sequencesinfragment3correspond
to the mutant regions ofts25 and ts616 (see
below).
ThelatemutantstslO, ts1260,and tsC as well
astheplaquemorphologymutant208 were res-cued by fragment 1 preferentially. Combining the resultsthat these latemutants can be res-cued with the HindlII-44% fragment, the
HpaII-1 fragment, and theHhaIIEcoRI-1
frag-ment,the location of themutant regionsof the
genome canbe placed between approximately
26.6and45.4 map unitsfrom theEco RI site.
Marker rescue with HhaI CR fragments. For furtheranalysis of the earlymutants, we measured the markerrescueability of two
frag-mentsproduced byHhaI cleavage of a polyoma
variant,CR. This variantlacks theHhaI
cleav-age site at 26.4 map unitsfrom theEcoRIsite
(M. Griffiths and C. Barry, unpublished 828
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.503.57.448.76.441.2]POLYOMA GENETIC MAP
Shalha
el-~
_ - 11h
HhO _
O0t
Hrin l
25 50
2 6 1 3
t
t
Hh al
tu±o11
ECO Rl 75
151
4 1817120T
EARLY RNA EARLY RNA
LATE RNA
tsa ts609
ts 25 ts48 ts 52 ts616 ts 697
- ts10
ts1260
tsc
[image:6.503.40.452.62.531.2]208
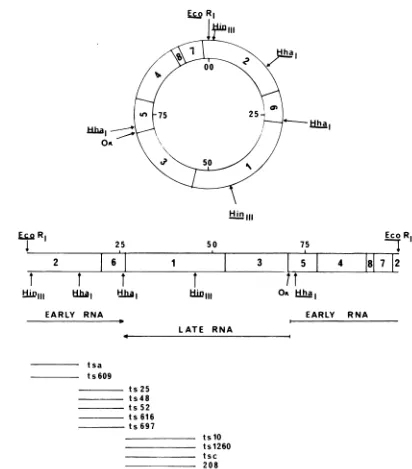
FIG. 2. PhysicalandgeneticmapofthepolyomaDNAgenome.At thetopisacircularphysicalmapofthe
polyomaDNAgenome(10, 11) showing thecleavagesitesofcertain restriction endonucleasesfrom
Esche-richiacoli (Eco RI), Haemophilus influenzaeD(HinIII), and H. haemolyticus(Hhal). The position of the
originofDNAreplicationisdenotedby (OR). Within the concentric circlesarethepositions ofthecleavage
sites ofa restriction endonuclease fromH.parainfluenzae (HpaII). Thearrangement ofthe eight HpaII
fragments, numbered in orderof decreasing size, is shown. Themapunits,with the Eco RI siteat00,are
marked in thecenterofthecircle;the 100mapunitscorrespondtoapproximately 5,200nucleotides. Below the
circularmapisalinearrepresentationofthesamephysicalmap.Theregionscorrespondingtoearlyand late
mRNAtranscriptsaredesignated by lines,thearrowsdesignatingthe directionof transcription (15). Finally
wehave denoted thepositions ofthemutantsitesoftsa,ts609, ts25, ts48, ts52, ts616,ts697,tslO,ts1260, tsC,
and 208 withrespecttothephysicalmap.
lr,
Ig
RI
VOL. 18, 1976 829
on November 10, 2019 by guest
http://jvi.asm.org/
TABLE 3. HhaI +EcoRI (39C/32C) x 100
Mutant
1 2 3 4
Early
tsa 0.2 1.0 7.0*a 1.0
ts25 0.5 1.7 4.6* 6.7*
ts616 0.12 1.5 7.8* 5.0*
Late
tslO 8.7* 2.4 1.4 1.1
ts1260 3.5* 0.3 0.1 <0. 1
tsC 8.5* 3.7 1.4 1.0
Plaque
mor-phology
208 57*W 20 <7 <7
Asteriskindicates themostpositive response. Dataforplaquemorphologymutant208 areexpressed
as(large plaque/small plaque)x 100.
data). Thus, HhaI cleavage of CR generates two fragments, one approximately 58% of the genome (designated HhaI CR-58%) and the other42%ofthe genome (designatedHhaI CR-42%). The results of marker rescue with the
HhaI CRfragmentsareshowninTable4.Some
oftheearly mutantswererescuedby HhaI CR-58% (e.g., ts25, ts48, ts52, ts697, and ts616), whereas others were rescuedby HhaI CR-52% (e.g, tsa and ts609). It is therefore possible to distinguish different mapping regions for early mutants by using these fragments.
Marker rescue byHhaI fragments. To fur-therlocate those earlymutants that mappedin the HhaICR-58% fragment, the marker rescue
abilityofHhaIfragments derived from theA-2
strainofpolyoma wastested. Three HhaI
frag-ments are produced from this strain: (i) a 46%
fragment, HhaI-46%; (ii) a 42% fragment, HhaI-42%; and (iii)a 12% fragment, HhaI-12% (10). To simplify the testing, we combined the
46 and 42% fragments and tested them
collec-tively, since our purpose was primarily to distinguish which region of the HhaI CR-58% region was responsible for marker rescue of certain earlymutants (see Table 4).
Table5presents the results of marker rescue ofearly mutants by either thecombined
HhaI-46% andHhaI-42% fragmentsortheHhaI-12%
fragment. Those early mutants thatpreviously wererescued by HhaI CR-58% (ts48, ts52, ts697, ts616, andts25; see Table 4) were rescued by the HhaI-12%fragmentofA-2. The early mutants thatpreviously were rescued by HhaI CR-46% (tsa andts609;seeTable4)were rescued by the combinedHhaI-46% andHhaI-42% fragments as expected.
Genotypic analysis of the viral DNA pro-duced by marker rescue. The marker rescue techniquepresumably involves only the
conver-sionof the ts region ofthe mutantgenome to
thewild-typeformandshould not affect regions
ofthemutantgenomeoutside theregionofthe
wild-type
fragment
used to rescue the mutant DNA. This hypothesis is testable since tsa DNAismissinganHaeHIcleavage site, which is present in the A-2wild-type
DNA. The tsa DNA lacks the HaeIII cleavage site betweenHaellI-6 andHaeIII-20 that is located at 28.0 map units (in HpaII-1), resulting in a slower
migration of tsaHaeIII-6 and the absence of
HaellI-20whencomparedwith theHaellI
frag-mentsofA-2 (Millerand Griffin,
unpublished
data).
Virusstocks
produced
from theexperimentofwild-type HpaII-2 fragmentrescueoftsaDNA
(seeTable 1)wereplaque assayedat39 C, and nine plaques were picked. The plaques were
grownat39C toproducevirusstocks,and32P_ labeled viral DNA was prepared from infec-tions with each virus stock. The 32P-labeled
DNAsweredigestedwith theHaeIII and
elec-trophoresed through 7% polyacrylamide
gels.
Nineoutofnineplaques pickedwereshownto have the same HaeIII cutting pattern as the
originaltsaDNA, and yet eachplaque wasno
longer temperature sensitive (results not
shown). This resultdemonstrates that marker
rescue involves only a limited region of the genomeand thatthose virusesplaquingat39C
(see Table 1) were most likely derived from modification of the tsa genome within the HpaH-2 region only.
DISCUSSION
A genetic map ofthe polyoma viral genome hasbeen constructed by using a markerrescue
TABLE 4. HhaICR fragments
(39C/32 C) x 100 Folddifference
Mutant (58%/42%or
58% 42% 42%/58%)
tsa 0.7 3.1* 4.4
ts25 13.8*a 1.1 12.5
ts616 5.5* 1.3 4.2
ts48 36.0* 13.0 3.0
ts52 3.1* 0.67 4.6
ts609 0.1 8.5* 85.0
ts697 1.0* 0.27 3.7
"Asteriskindicates themostpositiveresponse.
TABLE 5. HhaIfragments
(39C/32C) x 100 Folddifference
Mutant
~~~~~~[(46%
+42%)!46% +42% 12% (46% +42%)/
ts48 <0.014 6.15*a 439
ts52 0.75 10.6* 14.1
ts609 23.6* 2.5 9.4
ts697 <0.03 2.8* 93
"Asteriskindicates themostpositiveresponse.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.503.260.451.433.643.2]POLYOMA GENETIC MAP 831
technique. The map is diagrammatically pre-sented in Fig. 2. Since high backgrounds were noted in certain sets ofRENFs, we tested the ability of at least three different sets of RENFs to rescue each mutant. The results of each of thesesets were consistent, and the results are
summarizedinFig. 2.
Of the seven early mutants tested, two (tsa
and ts609)arerescued by fragmentscontaining the DNA regionbetween the HindIII site at 1.0 mapunitsand the HhaI site at 14.0 map units. The other five early mutants (ts25, ts616, ts48, ts52, and ts697) arerescuedbyfragments con-taining theDNA region between the HhaI site at 14.0mapunitsand the HhaI site at 26.4 map units. Thus theearly mutants show a cluster-ing effect in the DNA region between 1.0and 26.4 map units,whichcorresponds to the 3'-OH terminal half of the "early" RNA (15). These mutants are of interest since most ofthem have beendemonstrated to bedefective in transfor-mation attherestrictive temperature (4, 8).
Of the late mutants, tslO and ts1260 have
beenclassifiedintodifferentgroupsbygenetic
complementationtest (2-4). Thethird late mu-tant, tsC (Fried and Griffith, manuscript in preparation), efficiently complements early mutantsbutappears toshowpoor
complemen-tation with mutants of the other two late groups (W. Eckhart, personalcommunication)
andisthereforereminiscent of theBCgroupof
SV40. Our results indicate that ts10, ts1260, and tsC all map between 26.6 and 45.4 map units. This is the region specifiedby the 16S late stable mRNA that has been shown to
encode the majorcapsid proteinVP1 (26). Thus itwould appearthatthe complementation
ob-servedbetween tslO and ts1260 represents
in-tragenic complementation and not intergenic complementation. This finding is similar to
that observed for SV40, where the mutants from the twolate complementing groups (Band C)and thethirdlate group(BC,which
comple-ments early mutants but not other late
mu-tants)all mapinaregion of the SV40genome
that apparently encodesthe SV40 majorcapsid protein VP1 (17, 19, 22).
The plaque morphology characteristics of208 aredetermined by thesame regionof DNAin
which thelatetsmutantsts10,ts1260, andtsC map (26.6 to 45.4 map units). Hence the
mi-nute-plaque morphology of 208 is determined
bythe majorcapsid proteinVP1rather thanan
earlygeneoraltered sequenceatthe origin of DNA replication. Our results with HindIII
fragments have been confirmed by an
inde-pendent mappingtechnique (20) that involves
the in vitro recombination offragments from
large-plaqueA2andminute-plaque208parents
(Miller and Fried, unpublished experiments). Intheseexperiments, the hybrid DNA contain-ing the 56% HindHI fragment from large-plaque viral DNA and the 44%HindIII frag-ment from 208 DNA produced minute-plaque virus, whereas the hybrid DNA containing the 56% HindIII fragment from 208 and the 44%
HinduI
fragment from large-plaque virus pro-duced large-plaque virus upon infection of mousecells with the hybrid DNAs.The genetic organization of polyoma virus appears to be verysimilar to the genetic organi-zationof SV40determinedbyLai and Nathans (17). Mutants of both viruses show a distinct clustering effectinthesense that no mutations have been mapped inapproximately 55% of the genome, which comprises roughly half of the early region and half of the late region
sur-roundingtheorigin of DNAreplication. In the caseofpolyoma, we have yet to test two types of mutants, the host range mutants isolated by Benjamin (1) and the ts-3 mutant described by Eckhart and Dulbecco (5). This latter mutant
has similar properties to the class D mutants that Lai and Nathans have mapped in SV40
(17),
which laytowards the 5' terminus of the late RNA.Themarker rescue technique is a directand
relatively easy method for constructing a rough genetic map ofsmall, DNA-containing animal
viruses.Although more precisedeterminations
of the genetic map positions can be made by using smaller restriction endonuclease
frag-ments, such fragments are more difficult to
purify and would be time consuming to test. Moreprecise measurements of map locations of certain mutantsof SV40have beenprovidedby the
S,
nuclease technique ofShenketal. (25). TheS,
technique, however, can suffer from two majordisadvantages: (i) itis purely biochemi-cal and cannot relate base changes to pheno-typic differences; and (ii) the mapping proce-dure apparently cannot distinguish allsingle-basemismatchesand is therefore the method of choice for mappingdeletionmutationsonly.
We have studied certain aspects of the marker rescuetechnique ingreaterdetail. We
havedemonstrated that the region ofthe mu-tantgenome coveredbyawild-typerestriction endonuclease fragment is converted to wild-type, whereas sequencesinanother regionnot coveredby thewild-type fragment remain
un-changed. The cellular events involved in the marker rescue technique are presently being
studied.
Ourresults demonstratenotonlythe useful-ness of marker rescue for thegenetic mapping oftsandplaque morphology mutants of
poly-oma but alsothe usefulness ofthis method in VOL. 18, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
transferring genetic markers from one virus strain toanother.
ACKNOWLEDGMENTS
We wish tothank Beverley E. Cooke for her excellent technical assistance. We also thankBeverly E.Griffinfor helpand information on thephysical order of restriction endonuclease fragments during the course ofthiswork.
L.K.M. wassupported by Public Health Service Postdoc-toral Fellowship no. 1F22 CA 01198-01 from theNational Cancer Institute.
LITERATURE CITED
1. Benjamin, T. 1970. Host range mutants of polyoma virus.Proc. Natl. Acad. Sci. U.S.A. 67:394-399.
2. DiMayorca,G.,J. Callendar, G. Marin, and R.
Gior-dano. 1969. Temperature-sensitive mutants of poly-oma virus.Virology 38:126-133.
3. Eckhart, W. 1969. Complementation and transforma-tion by temperature-sensitive mutants of polyoma virus. Virology38:120-125.
4. Eckhart, W. 1974. Properties of temperature-sensitive mutants of polyoma virus.Cold Spring Harbor Symp. Quant. Biol. 39:37-40.
5. Eckhart, W., and R. Dulbecco. 1974. Properties of the ts3mutantof polyoma virus during lytic infection. Virology 60:359-369.
6. Francke, B., and W. Eckhart. 1973. Polyoma gene function required for viral DNA synthesis. Virology 55:127-135.
7. Fried, M. 1965. Isolation of temperature sensitive mu-tantsofpolyomavirus. Virology 25:669-671.
8. Fried,M. 1970.Characterization of a temperature
sen-sitive mutantofpolyomavirus.Virology 40:605-617. 9. Fried, M. 1974.Isolation and partial characterization of
differentdefectiveDNA moleculesderived from
poly-oma virus.J.Virol. 13:939-946.
10. Griffin, B. E., and M. Fried.1975. Amplification of a
specificregion of thepolyoma virus genome. Nature
(London)256:175-179.
11. Griffin,B.E., M.Fried, and A. Cowie. 1974. Polyoma
DNA: a physical map. Proc. Natl. Acad. Sci. U.S.A. 71:2077-2081.
12. Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369.
13. Hutchison, C. A., III, and M. H. Edgell.1971. Genetic assay for small fragments of bacteriophage 4X174 DNA. J. Virol.8:181-189.
14. Ishikawa, A., and G. Di Mayorca. 1971. Recombination between two temperature-sensitive mutants of
poly-oma virus, p. 294-299. InLepetitColloq.Biol.Med.,
The biology of oncogenic viruses, vol. 2.
North-HollandPublishers, Amsterdam.
15. Kamen, R., D. M. Lindstrom, H. Shure, andR. Old. 1974. Virus-specific RNA in cells productively
in-fectedortransformedby polyomavirus. ColdSpring
Harbor Symp. Quant. Biol. 39:187-198.
16. Lai,C-J., and D. Nathans. 1974.Mapping temperature-sensitive mutants ofSV 40: rescue of mutants by fragments ofviralDNA.Virology60:466-475. 17. Lai,C-J.,and D. Nathans. 1975. A map of
temperature-sensitive mutants ofSV40. Virology 66:70-81. 18. Mantei, N., H. W.Boyer, and H. M.Goodman. 1975.
Mapping SV40 mutants by construction ofpartial heterozygotes. J.Virol. 16:754-757.
19. May, E., H.Kopecka, and P.May.1975.Mapping the transcription site of the SV40-specific late 16S mRNA.Nucleic Acids Res. 2:1995-2005.
20. Miller, L. K., and M. Fried. 1976. In vitro construction ofinfectiouspolyomahybrid genomes. Nature
(Lon-don)259:598-601.
21. Miller, L. K.,and R. D. Wells. 1972. Properties of the
exonucleolyticactivities of the M.luteusDNA
polym-erase.J. Biol. Chem. 247:2667-2674.
22. Prives,C. L., H. Aviv, B. M. Paterson, B. E. Roberts,
S. Rozenblatt, M. Revel, and E. Winocour. 1974.
Cell-free translation of messenger RNA ofSV 40:
synthesis of the major capsid protein. Proc. Natl.
Acad. Sci. U.S.A. 71:302-306.
23. Radloff, R., W. Bauer, and J. Vinograd. 1967. A
dye-buoyantdensitymethod for thedetectionand
isola-tionofclosed circular DNAinHeLa cells. Proc.Natl.
Acad. Sci.U.S.A. 57:1514-1521.
24. Sharp,P. A., B. Sugden, and J.Sambrook. 1973.
Detec-tionof two restriction endonuclease activitiesin H.
parainfluenzae using analytical agarose-ethidium
bromideelectrophoresis. Biochemistry 12:3055-3063.
25. Shenk,T.E.,C.Rhodes,P. W. J. Rigby,andP.Berg.
1975. Biochemical method for mapping mutational alterationsinDNA withS, nuclease: thelocationof deletions and temperature-sensitive mutations in
SV40.Proc. Natl.Acad. Sci. U.S.A. 72:989-993.
26. Smith, A. E., S. T. Bayley, W. F. Mangel, H. Shure, T. Wheeler, and R. I. Kamen. 1975. Characterization and translation ofpolyoma virus messenger RNAs, p. 151-164. In FEBS Symposium. North-Holland, Amsterdam.
27. Smith, H. O., and K. W. Wilcox. 1970. A restriction enzyme from H. influenzae. I. Puridication and gen-eralproperties. J. Mol. Biol. 51:379-391.
28. Weisbeek, P. J., and J. H. Van DePol. 1970. Biological activity of4X174replicative formDNA fragments. Biochim.Biophys. Acta 224:328-338.
on November 10, 2019 by guest
http://jvi.asm.org/