YANG, SOPHIA. Arbovirus Structure and Interaction with Host Cells. (Under the direction of Dr. Dennis Brown).
The first study presented investigates the role of Sindbis E1 transmembrane domain on virus assembly. The Sindbis virion is comprised of two proteins shells formed by three structural proteins. Capsid (C) proteins forms the inner shell and encompasses the viral RNA genome. The outer glycoprotein coat is formed by 240 copies each of the E1 and E2 proteins that form trimers of heterodimers to make the characteristic “spikey” outer structure. E1-E2 transmembrane
domain (TMD) interactions are known to be important for the overall stability of a mature virion. A previous study has found mutations within the E2 TMD affect viral assembly and infectivity. The study concluded that the location of the amino acid deletions on the hydrophobic face of the TMD helix and toward the c-terminal end had a great effect on viral production. It was found that large deletions within the E2 TMD up to 10 amino acids were tolerated. To further
investigate the role of the glycoproteins’ TMD on viral assembly and infectivity, deletions of the E1 TMD were constructed. In contrast to the E2 TMD, large deletions within the E1 TMD and deletions made on the hydrophilic face of the helix were not tolerated. These results suggest specific functions of the different regions in the E1 TMD and its interaction with the E2 TMD indicating that E1 stabilizes the E1/E2 spike complex.
strains share 96% sequence identity and analysis of the sequences have not identified the cause for increased virus pathogenesis. We hypothesize that the differences lie in how the Zika strains regulate microRNA expression thereby leading to a difference within host cell immune response. It this study, we chose miR-146a as our target as it if found during the course of many RNA viral infections and is known to regulate the toll-like receptor innate immune response. Human
monocytes and MФs were infected with the AF (MR766) and AS (PRVABC59) Zika strains.
Using miR-146a as our target regulator, the expression of select genes associated with
by Sophia Yang
A dissertation submitted to the Graduate Faculty of North Carolina State University
in partial fulfillment of the requirements for the degree of
Doctor of Philosophy
Biochemistry
Raleigh, North Carolina 2019
APPROVED BY:
_______________________________ _______________________________ Dennis Brown Raquel Hernandez
Committee Chair
_______________________________ _______________________________ Michael Goshe Flora Meilleur
_______________________________
ii DEDICATION
iii BIOGRAPHY
Sophia was born in Sacramento, California and raised in Charlotte, North Carolina. She found her calling in scientific research during her undergraduate career at North Carolina State University. During this time, Sophia did her undergraduate research in Trino’s lab studying senescence in Arabidopsis thaliana plants infected with Geminivirus. Sophia started the
Biochemistry PhD program at NCSU in Dr. Bob Rose’s lab and is finishing the program in Dr. Dennis Brown’s lab.
iv ACKNOWLEDGMENTS
My list of people to thank for all of their contributions to my success is endless, but there are a few that deserve special mention:
First and foremost; Drs. Dennis Brown and Raquel Hernandez. Thank you for allowing me to complete my graduate career in your lab. You have challenged me to think more critically, allowed me to formulate my own experiments and given me the opportunity to work with some truly amazing people. Your guidance and friendship have made me the scientist I am today. To the rest of my graduate committee both past and present: Drs. Bob Rose, Michael Goshe, Flora Meilleur, Frank Scholle, and Rodolphe Barrangou. Thank you for your guidance and
encouragement throughout my education.
My lab mates, both past and present: Dr. Davis Ferreira, Dr. Alex Breuer, Susan May, Dr. Ryan Schuchman, Dr. Joe Magliocca, Dr. Greg Burhman, Dr. Paul Enrique and Luke Dillard. It has been a pleasure working with each of you. You all are the best lab mates!
Dr. Trino Ascencio-Ibanez, you are the reason why I chose to go into scientific research and I am forever grateful. Thank you for being my teacher, mentor and friend.
Dr. Paul Swartz, thank you for helping keep me sane throughout my graduate career. I have enjoyed our conversations and your blunt advice.
v Last, but not least, I would like to thank Daniel O’Brien. Thank you for helping me get back up when I think I’ve hit rock bottom, for being patient with my long days and weekends at
vi TABLE OF CONTENTS
LIST OF TABLES ... vii
LIST OF FIGURES ... viii
Chapter 1: Deletions within Sindbis Virus E1 Glycoprotein Transmembrane Domain and their Role in Virus Assembly ... 1
1.1: Abstract ... 2
1.2: Introduction ... 3
1.3: Results ... 13
1.4: Discussion ... 26
1.5: Methods ... 31
1.6: References ... 35
Chapter 2: Role of miR-146a on Zika Virus Pathophysiology ... 44
2.1: Abstract ... 45
2.2: Introduction ... 47
2.3: Results ... 54
2.4: Discussion ... 63
2.5: Summary ... 72
2.6: Future Direction ... 73
2.5: Methods ... 74
2.6: References ... 84
APPENDICES ... 93
Appendix A: Table of Tested Yeast 1 Hybrid Gene Hits ... 94
vii LIST OF TABLES
Table 2.1 Primer sequences used for psiCHECK-2 and pAbAi vector mutations. Primers were designed using Primer BLAST (https://www.ncbi.nlm.nih.gov
/tools/primer-blast/). ... 77 Table 2.2 Primer sequences used for qRT-PCR. Primers were designed using Primer
viii LIST OF FIGURES
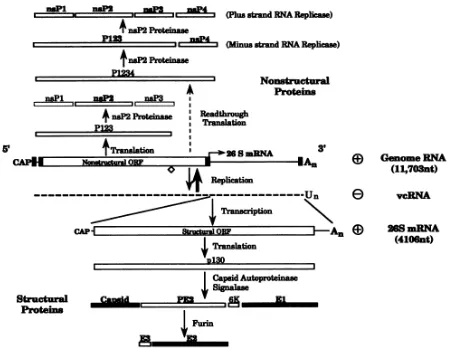
Figure 1.1 Schematic of Sindbis virus genome (from Strauss, 1994, with
permission). The 49S genomic RNA is illustrated in the center and is labeled as the positive Genome RNA. The non-structural proteins are translated directly from the 49S open reading frame. Structural proteins are translated and processed from the 26S subgenomic mRNA, which is transcribed from the negative strand complement of the genomic RNA,
vcRNA ... 6 Figure 1.2 A schematic representation of the organization of the Sindbis virus
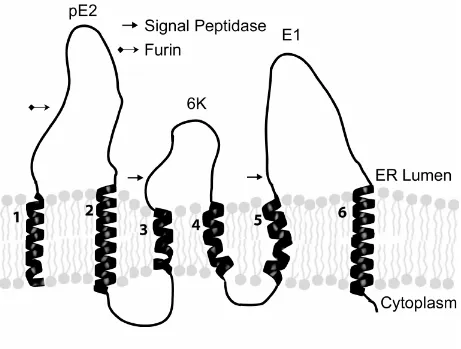
structural proteins in the membrane of the Endoplasmic Reticulum (from Whitehurst et al., 2006, with permission). The integrated type 2
polyprotein has six transmembrane domains labeled 1 to 6. The 6K protein is eliminated from the developing spike complex after processing by signal peptidase. The N-terminal region of PE2 is removed in the Golgi by Furin protease. Of the remaining three domains that are incorporated into virus, domain 3 is withdrawn from the membrane and attaches to the nucleocapsid, and domains 2 and 6 are membrane
anchors ... 7 Figure 1.3.1 E2 Transmembrane domain mutants (from Whitehurst et al., 2006, with
permission ). A) Helical wheel representation of the E2 transmembrane domain. The arrows denote the single amino acid deletions which cover the entire face of the helix. The hydrophobic and hydrophilic faces have been labelled. Mutants that produced viral titers of 10^6 pfu/mL are boxed while the other mutants produced 10^8 pfu/mL. B) Sequences of the E2 mutants where (-) denotes the location of amino acid deletion. The resulting number of amino acids left in the domain are listed to the
right ... 9 Figure 1.3.2 Mutant E2 titer and relative infectivity (from Whitehurst et al., 2006,
with permission). A) Viral titer of E2 mutants truncated to 25 or 18 amino acids. The arrow at the bottom denotes the relative location of the TM25 deletions from the N- to C-terminus. SVHR represents wildtype Sindbis. B) Relative infectivity of the truncated E2 mutants. Mutants that produced higher titers had a lower particle-to-pfu ratio than mutants
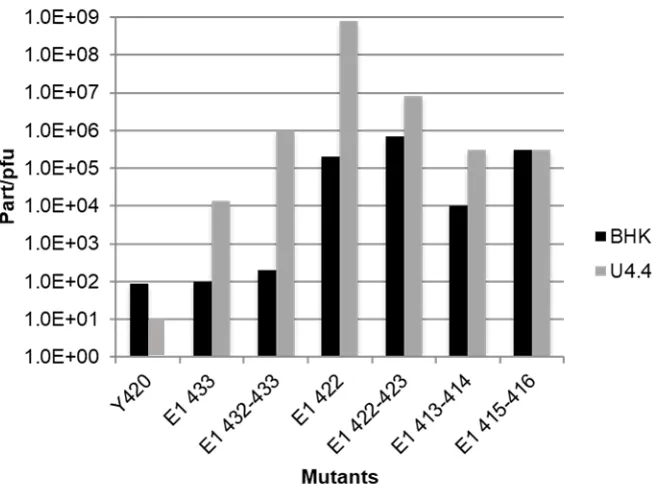
that produced lower viral titers ... 10 Figure 1.4 Systematic truncation of the E2 transmembrane domain and
ix Black bars indicated BHK, mammalian, cells and grey bars indicate
U4.4, insect, cells ... 11 Figure 1.5 A) Helical wheel representation of the E1 transmembrane domain.
Outlined residues shows the amino acids that were deleted. The mutations were made to encompass the entire helical face. The central arrow indicates hydrophobic face. B) Sequences of the mutants constructed showing the amino acids deleted (shown in grey and underlined). The sequence between E1 residues 407 and 435 were chosen as residues likely found within the TMD of E1. The mutant viruses are named for the aa number of the deleted residues. C) Virus infectivity of the mutants compared to wild type virus. Each of the mutants constructed was transfected into BHK and U4.4 cells then subsequently titered by plaque assay on BHK cells. Data represents the average of 2 independent experiments with three technical replicates
each ... 15 Figure 1.6 A) Mutants of the E1 transmembrane domain were generated by
truncating the E1 transmembrane domain from 28 amino acids to 14 or 18 amino acids. B) BHK and U4.4 cells were transfected with the mutants and the viral titer was determined by plaque assay on BHK cells. Wild type viral titer was compared to viral infectivity of each of the mutants. Compared to the wild type virus, each of the mutants lost titer by 3 to 7 logs or produced no infectious virus. Data represents the average of two independent experiments with three technical replicates
each. The standard deviation of the averages is also plotted ... 18 Figure 1.7 Synthesis of viral proteins from non-infectious mutants. As shown in
lanes 3 (E1Δ 420-433), 4 (E1Δ 411-428), and 5 (E1Δ416-433), viral structural proteins are synthesized and processed in BHK cells which did not generate any infectious virus particles. The accumulation of pE2 in these cells suggests that formation of the heterodimer of pE2 with the deleted E1 mutants may alter the conformation of the complex in a way
which affects furin cleavage ... 21 Figure 1.8 The particle-to-pfu ratios of the E1 mutants compared to wild type virus.
E1Δ 422, E1Δ 422-423, E1Δ413-414, produced ~ 105.5 ,106 and 105 orders of magnitude of non-infectious virus respectively in BHK cells. Mutants E1Δ433 and E1Δ432-433 produced comparable amounts of infectious virus to Y420 even though these mutants produced lower viral
x Figure 1.9 Ultrastructure of BHK cells transfected with the mutated E1 strains. (A)
BHK cell transfected by non-viral RNA. (B) BHK cell transfected with Y420 strain (wild type control). The arrow points at a cluster of
assembled nucleocapsids while the arrowheads point to viruses budding from the plasma membrane. (C) BHK cell transfected with E1Δ 420-433. Arrows show nucleocapsid modified membranes which are
accumulating internally. Note that budding particles are not visible at the
cell surface. (N denotes the nucleus.) Magnification bars are 300 nm ... 25 Figure 2.1 Phylogenetic tree of Zika virus comparing isolates found from GenBank
and outlines the African and Asian lineages. The viruses used for this study are circled in red with MR766 stemming from the African lineage and PRVABC59 from the Asian linage. P6-70 was isolated in Malaysia, the first Zika isolate to migrate from Africa into Asia. The scale bar indicates the number of nucleotide substitutions per site. (Image adapted
from Lanciotti et al., 2016b) ... 48 Figure 2.2 Zika Genome Organization. A) Single-stranded positive sense RNA
genome is about 11kb and can be divided into structural and non-structural coding regions. The entire genome is translated into one large polyprotein in the cytoplasm of the infected cell. The products of the proteolytic processing cascade are shown in the boxes below the genome. B) Topology of the polyprotein in the Endoplasmic Reticulum
membrane. (image from Shi & Gao, 2017, with permission) ... 50 Figure 2.3 Monocytes (-PMA) and MФ (+PMA) were infected with MR766 or
PRVABC59. Viral titer was determined from samples collected intermittently between 2- and 56-hours post-infection. Cells infected with the AS strain of ZIKV were shown to produce 300-fold more infectious virus 8 hours post-infection than those infected with the AF strain. AS infected cells continued to produce higher titers of virus throughout the infection time course, expressing distinctly faster and
longer kinetics. Data from two independent experiments are displayed ... 55 Figure 2.4 Endogenous miR-146a levels in monocytes (-PMA) or macrophages
(+PMA) infected with either MR766 or PRVABC59 at (A) 2, (B) 12 and (C) 24 hours post-infection. Levels were normalized against the
expression levels found in uninfected monocytes or macrophages. The untreated monocytes have higher levels of miR-146a than the treated, differentiated THP-1 cells. In contrast to PRVABC59, cells infected with MR766 have greater levels of the miRNA than those infected with PRVABC59. Data was collected from three technical replicates.
xi Figure 2.5 Expression levels of yeast-1 hybrid (upstream) gene hits in THP-1 cells
that were (A) untreated and (B) treated with PMA. Viral samples were collected from cells infected with either MR766 or PRVABC59 12 hours post-infection. Results are relative to the expression levels in uninfected monocytes and macrophages. Data points represent three
technical replicates. Standard deviation error bars are plotted ... 59 Figure 2.6 Expression levels of luciferase (downstream) gene hits in THP-1 cells
that were (A) untreated and (B) treated with PMA. Viral samples were collected from cells infected with either MR766 or PRVABC59 12 hours post-infection. Results are relative to the expression levels in uninfected monocytes and macrophages. Data points represent three
technical replicates. Standard deviation error bars are plotted ... 60 Figure 2.7 Expression levels of genes found to be directly associated with known
networks. Monocytes (-PMA) and macrophages (+PMA) were infected with either MR766 or PRVABC59. Samples were collected (A) 2, (B) 12 and (C) 24 hours post-infection. Results are relative to the expression levels in uninfected monocytes and macrophages. Data points represent the results of three technical replicates. Standard deviation error bars are
plotted ... 62 Figure 2.8 IL-6 signaling pathway. IL-6 binds to membrane receptors that contain
gp 130 and activates the JAK/STAT pathway and the MAPK cascade. The expression levels of IL-6 and PIK3CB, the catalytic isomer of p110, were evaluated in this study and are circled in red. This figure was originally published in (Heinrich, Behrmann, Haan, Hermanns, &
Uller-newen, 2003). Shown with permission ... 66 Figure 2.9 NFĸβ signaling pathway. TRAF6, IRAK1 and PIK3CB (PI3K subunit)
are all shown to be upregulated 4-8-fold in monocyte and macrophage cells infected with PRVABC59 (refer to Figure 2.6). These genes, circled in red, are required for NFĸβ production. These proteins function as dimeric transcription factors that regulate the expression of genes that influence cell survival and innate immune response. Image originally found at www.novusbio.com/NFĸβpathway (Novus Biologicals, 2019)
and is used with permission ... 69 Figure 2.10 Toll-like receptor signaling pathway schematic. MyD88-dependent
pathways utilize IRAK1 and TRAF6 proteins, downstream effectors of miR-146a circled in red, to ultimately produce inflammatory cytokines. Image originally found in (Guo, Zhang, & Jia, 2018). Reproduced with
1 CHAPTER 1
Deletions within Sindbis Virus E1 Glycoprotein Transmembrane Domain and their Role in Virus Assembly
Sophia Yang, Mariana Ribeiro, Gongbo Wang, Joe Kononchik, Ricardo Vancini, Dennis T. Brownand Raquel Hernandez
2 1.1 Abstract
The Sindbis virion contains an RNA dense core and a host-derived membrane bilayer that is sandwiched between two protein shells. The shells are comprised of 240 copies of each of the three structural proteins E1, E2 and Capsid (C) in a 1:1:1 stoichiometric ratio and the virion is arranged in a T=4 quasi-icosahedral conformation. Both E1 and E2 have transmembrane
3 1.2 Introduction
Sindbis virus (SINV) is an Alphavirus with a positive sense, single stranded RNA genome and is transmitted in nature between birds and mammals by blood-feeding mosquitos. Currently, there are more than 40 members of the Alphavirus genus which has a worldwide geographical distribution and it is found on every continent (La Linn et al., 2002). Humans are an incidental host of these viruses. The alphavirus genomes share about 45% sequence identity in the structural proteins and about 60% in the nonstructural proteins (J. H. Strauss & Strauss, 1994). Alphaviruses are grouped by geographic distribution into Old World and New World viruses. The pathogenic alphaviruses produce disease that generally occurs in two forms, febrile with accompanying malaise such as in the New World alphaviruses (concentrated in the
Americas) and include the equine encephalitis strains (eastern, western, and Venezuelan equine encephalitis viruses, EEE, WEE and VEE, respectively) which are pathogenic in humans, horses and some bird species. Old World viruses are commonly associated with fever, rash and
rheumatic disease in humans and include chikungunya virus (CHIKV), Mayaro (MAYV) and Ross River viruses (RRV). SINV and Semliki Forest virus (SFV) constitute non-pathogenic alphaviruses and are well characterized. SINV was first discovered in 1952 during an outbreak in Cairo, Egypt (Lwande et al., 2015; Taylor et al., 1953). Since then, there have been several outbreaks of Alphavirus. One of the most notable examples was the CHIKV outbreak in 2005 where 40% of the La Reunion population was infected resulting in 250 fatalities (Bessaud et al., 2006; Josseran et al., 2006).
4 composed of two nested protein shells arranged in an icosahedral T=4 lattice (Paredes et al., 1993). The inner protein shell of the virus is comprised of the RNA viral genome enclosed by a C protein shell. The outer shell is made of E1 and E2 glycoproteins that are arranged in dimers of heterodimers which form the 80 trimers on the virion surface, giving it a “spikey” appearance. (Anthony & Brown, 1991; Brown, Wan, & Kielian, 2018; Carleton, Lee, Mulvey, & Brown, 1997; Pletnev et al., 2001). A host-derived lipid bilayer is sandwiched between the two shells incorporating the transmembrane (TM) domains of the E1 and E2 proteins which span the membrane (Liljeström & Garoff, 1991; Rice et al., 1982; Strauss et al., 2002). E2 has an
extended 33 amino acid tail that emerges from the transmembrane domain into the cytosol of the host cell. It is to this tail that the C protein binds, making the viral structure symmetrical and rigid (Ferreira et al., 2003; Hernandez et al., 2003).
The genomic RNA of SINV is about 11.7kb with a 5’ m7Gcap and 3’ poly-A tail. The
genome is divided into two regions, structural and nonstructural domains, which encode for the structural and nonstructural viral proteins (Figure 1.1). The nonstructural proteins are translated directly from the genomic plus sense 49S RNA and cleaved to produce nsP1-4 and other
5 in the cytoplasmic ribosome, exposing a signal sequence at the amino terminus of the
polypeptide (Ferreira et al., 2003; Liljeström & Garoff, 1991) This signal sequence directs the polypeptide to be inserted in the endoplasmic reticulum membrane where the newly synthesized envelope glycoproteins are translocated. The transmembrane polyprotein has 6 membrane spanning domains (Figure 1.2) and is processed into PE2 (E3/E2), 6K/TF, and E1 by signal peptidase (Liljeström & Garoff, 1991). The 6K protein has two transmembrane domains which are shown as domains 4 and 5 in Figure 1.2. 6K is released from the ER membrane following signal peptidase cleavage and is present at very low levels in mature virus (Mukhopadhyay et al., 2006). 6k also aids in the correct integration of E1 into the membrane as translocation proceeds (Liljeström & Garoff, 1991). During the process of maturation, E1 forms heterotrimers with PE2 (Carleton et al., 1997; Mulvey & Brown, 1996). As the complex migrates to the cell surface, furin protease, which is found within the trans Golgi network (Moehring et al., 1993), processes PE2 to form the E2 and E3 proteins. However, E3 is not present in the mature virus and is shown as domain 1 in Figure 1.2. The E1 and E2 proteins dimers convert to their final trimeric
7 Figure 1.2: A schematic representation of the organization of the Sindbis virus structural
8 The E1 and E2 glycoproteins are the only proteins that remain membrane associated in mature SINV. Although the specific amino acid sequence for the transmembrane domains of these two proteins is not conserved among alphaviruses, the length (26-28 amino acids) and hydrophobic characteristics are conserved. Previous studies have investigated the E2
transmembrane domain and its role in viral assembly and infectivity (Whitehurst et al., 2006). This was done by truncating the transmembrane region with single to multiple amino acid deletions that encompass the entire helical face (Figure 1.3.1a). It was found that the location of the deletion along the vertical axis of the transmembrane domain is important for normal virus maturation (Whitehurst et al., 2006). Single amino acid deletions toward the N-terminus of the TMD are more tolerated than those made toward the C-terminus (Figure 1.3.2a). Mutants that had higher viral titers had more infectious particles than mutants that produced lower viral titers. Additionally, the particle-to-pfu ratio (number of physical particles/ numbers of infectious particles) of mutants with higher virus titer have a lower particle-to-pfu ratios than mutants that had low titers (Figure 1.3.2b) suggesting that the defect conferred by the deletion affected structural stability and thus infection. Furthermore, deletions on the hydrophobic face of the E2 TMD helix have a greater impact on normal virus growth than those made on the hydrophilic face. Large deletions up to 10 amino acids in U4.4 mosquito cell- grown and 8 amino acids in virus grown in BHK cells were tolerated, producing viral titers comparable to those of wildtype virus (Figure 1.4). Although these deletions truncated the TM to 16-18 amino acids long, the domain was still able to span the lipid bilayer. It was concluded that the interaction between the E1 and E2 transmembrane domains was occurring within the cytoplasmic half of the E2
9 Figure 1.3.1: E2 Transmembrane domain mutants (from Whitehurst et al., 2006, with
10 Figure 1.3.2: Mutant E2 titer and relative infectivity (from Whitehurst et al., 2006, with
permission). A) Viral titer of E2 mutants truncated to 25 or 18 amino acids. The arrow at the bottom denotes the relative location of the TM25 deletions from the N- to C-terminus. SVHR represents wildtype Sindbis. B) Relative infectivity of the truncated E2 mutants. Mutants that produced higher titers had a lower particle-to-pfu ratio than mutants that produced lower viral titers.
A
11 Figure 1.4: Systematic truncation of the E2 transmembrane domain and corresponding viral titer (from Sinodis et al., 2003, with permission). A) Sequences of the E2 mutants. Amino acids in bold are those that were deleted for the mutation. The remaining number of amino acids left in the domain are listed to the right of the sequence. B) Viral titer of E2 mutants. Y420 represents wild type. Large deletions up to 10 amino acids in U4.4 (TM16) and 8 amino acids in BHK (TM18) are tolerated. Black bars indicated BHK, mammalian, cells and grey bars indicate U4.4, insect, cells.
A
12 To determine how the E1 TMD is interacting with the E2 TMD, a mutagenesis study similar to the one done for the E2 protein was conducted. Single and double amino acid deletions were made to transverse the helix from the N- to C- terminus and around the helical face (Figure 1.5a). Unlike the E2 TMD, deletions on the hydrophilic face of the E1 TMD helix were not tolerated and resulted in a 4-log loss in viral titer (Figure 1.5c). Shortening the E2
13 1.3 Results
1.3.1 Virus production from E1 TMD deletion mutants.
Previous studies found that the deletion of a single methionine at M379 toward the carboxyl terminus of the E2 TMD dropped virus titer by four orders of magnitude (Hernandez et al., 2003). Additional single deletions near the carboxyl terminus of the E2 TMD A385 and V386 were found to have a dramatic negative impact on virus infectivity (Whitehurst et al., 2006). Thus, it was found that one amino acid deletion could dramatically influence the titer of the virus. It was also apparent that the E2 TMD could withstand large deletions without deleterious effects to the virus titer. Additionally, E2 TMD could tolerate single amino acid deletions if these were located toward the amino terminus of the domain. It was observed that increasing deletions could restore the function of the E2 TMD probably due to the restoration of the helix toward the complete rotation established by 18 amino acids within the TMD. Based on these observations of the E2 TMD, a similar strategy was used to make deletion mutations in the E1 TMD (Whitehurst et al., 2006).
14 Based on these observations of the E2 TMD, deletion mutations were made in the E1 TMD. Methionine at position 433 in the carboxyl region of E1 TMD, a position also found to be important for virus infectivity in Sindbis-Ross River (RR) virus chimeras (Kim et al., 2002) was deleted in the mutant E1Δ433 (Fig. 1.5b). The titer of this mutant was found to be 100-fold lower than wild type virus from both mammalian and insect host cells suggesting that the carboxyl terminus of E1 is involved in virus assembly or infectivity (Fig. 1.5c). When both methionines at the carboxyl terminus of the E1 TMD were deleted in the mutant E1Δ432-433, the virus titers
16 Considering that the MM motif in E2 TMD is centrally located, the consequences of deletion of the centrally located duplicate isoleucines of the E1 TMD were investigated. There are two isoleucines (I422 and I423) centrally located in the E1 TMD domain; their functions were investigated in single and double deletions. The E1 isoleucine 422 was deleted (E1Δ422)
and produced a dramatic drop in titer of 1000-fold from both host cells (Fig. 1.5c). Deleting both isoleucines (E1Δ422-423) did not result in a host-restricted phenotype, by contrast, this mutation
allowed the virus containing this additional isoleucine deletion to regain virus production by two orders of magnitude (108 pfu/mL) over the single mutant in both host cells. (106 to 108 pfu/mL, Fig. 1.5c). From this result, we can conclude that E1 I422 is essential for virus production in the context of I423 but that removal of the isoleucine at position 423 can compensate for its absence. Thus, this result also appears to be a positional effect with respect to the amino and carboxyl ends of this domain resulting in G at position 422 (Lopez et al., 1994; Sjoberg & Garoff, 2003; Strauss et al., 2002).
Deleting amino acids at the proximal and distal regions of the E2 TMD resulted in differential virus infectivity. It was found that the least affected region of the E2 helix was toward the amino terminus (Whitehurst et al., 2006). To address the question if this may also be the case in E1, two adjacent amino acids near the E1 amino terminus were deleted (413L-414F and 415G-416G, Fig. 1.5b). The mutant, E1Δ413L-414F, has a titer which is one order of magnitude lower than the wild type when produced from both host cell types (109 pfu/mL). Loss of titer was also displayed by the mutant E1Δ415G-416G showing a greater effect on virus titer
17 membrane, this observation suggests that more than one function is being affected by these E1 deletions. Some interactions may involve the specific geometry of the E1 TMD which may be disrupted, in addition to physical interactions with the E2 TMD as predicted by structural studies. The remaining 26 amino acids should theoretically be able to span the membrane of both hosts. While this deletion does not have the more drastic effects of some of the others in the E1 TMD, the observed loss of titer in all the mutants which produced virus could be the result of loss of infectivity from instability or defective assembly.
Large deletions of E1 TMD were also constructed, truncating the domain to 10, 14 and 18 amino acids. These deletions were made near both the amino terminus (E1428, Δ411-420, Δ410-423 and Δ407-416) and carboxyl terminus (Δ416-433, E1Δ420-433, Δ421-430, Δ425-434). Unlike the E2 TMD deletion mutants that only required 10 residues to produce
functional virus in mosquito and mammalian cells (107 pfu/mL, Figure 1.4B), mutants with large deletions in E1 TMD produced little to no infectious virus from either host (Figure 1.6).
19 1.3.2 Viral protein expression in the E1 TMD lethal mutants
The mutants E1 Δ420-433, Δ411-428, and Δ416-433 have 14, 14 and 18 amino acids
deleted, respectively (Fig.1.6a). These are lethal mutants that did not produce any infectious virus from either host. When the same number of amino acids were deleted from the E2 TMD however, virus was still produced (Hernandez et al., 2003). Analysis of the phenotypes of the E1 TMD lethal mutants began with assays of viral structural protein synthesis, nucleocapsid assembly and budding.
To assay viral protein synthesis, BHK cells were transfected with each mutant viral RNA and protein was labeled metabolically with 35S. Viral protein was isolated from the cellular proteins using immunoprecipitation. Viral protein was analyzed with SDS-PAGE as shown in Figure 1.7. Our results show that protein expression is not impaired by the E1 transmembrane deletions because the structural proteins are being synthesized in the expected ratios. However, in wild type virus, the band of pE2, the E2 precursor, is not as intense as E1/E2 band, while in the mutants, the intensity of the two bands is about the same. This suggests that the maturation process of pE2 to E2 is not as efficient in these mutants.
The possibility that the lethal E1 mutants were defective in the processing steps prior to assembly was considered. For these deletion mutants, it was of interest to determine if they inserted the polyprotein precursor into the membrane and were processed correctly. Our results show that protein expression is not impaired by the mutations in E1 Δ420-433, Δ411-428, and Δ416-433 mutations because the structural proteins are being synthesized in the expected ratios
22 1.3.3 Relative Infectivity of the E1 TMD deletion mutants.
26 1.4 Discussion
Sindbis virus structural proteins are originally synthesized as a polyprotein in the sequence C-PE2 (E3-E2)-6K-E1. E1 and E2 are both integrated into the endoplasmic reticulum membrane during the process of polyprotein synthesis, after which enzymatic cleavages separate them. E1 and E2 form trimers of heterodimers in the ER prior to export to the Golgi. The co-expression of E2 is required for trafficking of E1 from the ER. Therefore, during protein maturation and assembly of the virus, these proteins perform their functions as metastable oligomers requiring lateral associations (Sjoberg & Garoff, 2003). Biochemical and genetic studies using chimeras of Ross River and Sindbis viruses and substitution mutations in Semliki forest virus as well as structural evidence suggest that the TMDs of E1 and E2 interact within the viral membrane.
27 length the helix makes five turns and contains residues equally distributed throughout the helical surface. In the E2 TMD the location of an eight amino acid deletion was tolerated at the amino but not the carboxyl terminus suggesting that E2 interacted with E1 toward the carboxyl terminus. The second aspect was the influence of the diverse biochemical lipid environments imposed by the insect or mammalian environments which enabled the expression of the E2 host range mutants. Mammalian membranes, which are thicker, more viscous and contain more cholesterol than insect cell membranes in culture, were not tolerant of the 9 and 10 amino acid deletions (Hernandez et al., 2005). It was hypothesized that the ability of E2 to tolerate such large mutations may result from a stabilizing effect of the E1 TMD or lateral interactions.
28 of the virus to undergo budding even though nucleocapsids were able to associate with the glycoprotein modified membrane. These observations suggest that these deletions disrupt the E1 lateral associations required for virus envelopment.
A more complicated phenotype to explain is the host range characteristic of E1Δ432-433,
29 Analysis of the hydrophobicity of the E1 and E2 TMDs by helical wheel analysis in Heliquest and Protean shows that E1 has a more hydrophobic nature than E2. These predictions suggest that E1 and E2 are not typical amphipathic helices with clear hydrophobic and hydrophilic surfaces. Our previous genetic analysis of the effects of amino acid deletions in E2 which affected virus titer are found in the more hydrophobic face of the helix with the exception of A385 (shown in Fig. 1.7) which is in agreement with other genetic studies of SIN and RR chimeras. When SINV E1 was investigated by deletion analysis, mutations affecting titer also mapped primarily to a single face of E1 but these were found to cluster toward the more hydrophilic face. This was also unexpected because it was expected that the two adjacent faces of E1 and E2 would interact. However, this was also the case in the E1 modeling of the reconstructions of SINV and VEEV referred to above. Another feature of these sequences is that SINV E2 TM18 deletes 8 consecutive amino acids but retains full infectivity. This is also interesting because E1 and E2 are modeled as coiled coils. Because these domains are in the membrane bilayer we will assume, for this discussion, that each face of these helices is defined by amino acids within 180 ° of the first amino acid in the hydrophobic face calculated by Heliquest (Figures 1.3.1A and 1.5A). The sequence of SINV TM18 365 VYTIL 370 AVASA 375
TVAMM 380 IGVTV 385 AVLCA 390 C (underlined sequence deleted) the helix sequence
does not come back into the original register until A389 (Fig. 1.8) This implies that amino acids known to affect titer, with the exception of I380, (replaces M379) relocate to the more hydrophilic face (Figure 1.3.1A).
31 1.5 Methods
1.5.1 Cell Culture
BHK-21, baby hamster kidney, cells were cultured in minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 5% tryptose phosphate both, 2mM glutamine, 50 µg/mL gentamicin and 10 mM HEPES (pH 7.4). Newly thawed cells may be passaged up to 30 times. Cells should be incubated at 37°C with 5% CO2 and split at a 1:3 ratio daily. Incubation at 28°C for two to three days is permissible, but must be split at least once to recover to normal growth patterns before use in experiments. U4.4, Aedes albopictus – mosquito, cells were cultured in minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 5% tryptose phosphate broth, 2 mM glutamine, 10 mM HEPES (pH7.4) and 7.5% sodium
bicarbonate. Newly thawed cells may be passaged up to 30 times without change in cell viability. Cells should be grown at 28 with 5% CO2 and split at a 1:3 every 4-5 days.
1.5.2 Sindbis virus E1 Mutant Construction
The TM mutants were made by QuikChange site-directed mutagenesis. The template cDNA used was Toto1101 (Rice et al., 1987). Specific primers were made for each of the TM mutants to delete the desired regions as shown in Figure 1.6A. The following primer pairs were used:
E1Δ411-420_Fwd: 5'-ctcaaaaacatcatggagttggctgttaattataggacttatgatttttg-3'
E1Δ411-420_Rev: 5'-caaaaatcataagtcctataattaacagccaactccatgatgtttttgag-3'
E1Δ421-430_Fwd: 5'-cggcgcctcgtcgctaagcatgatgctg-3'
32 E1Δ410-423_Fwd: 5'-ctcaaaaacatcatggagttggggacttatgatttttgcttgca-3'
E1Δ410-423_Rev: 5'-tgcaagcaaaaatcataagtccccaactccatgatgtttttgag-3'
E1Δ425-434_Fwd: 5'-catcttcgtgtgctagttcctataattaatagcgacgaggcg-3'
E1Δ425-434_Rev: 5'-cgcctcgtcgctattaattataggaactagcacacgaagatg-3'
E1Δ407-416_Fwd: 5'-gccgccatctcaaaaacatcagcctcgtcgct-3'
E1Δ407-416_Rev: 5'-agcgacgaggctgatgtttttgagatggcggc-3'
The QuikChange mutants were made using Pfu Turbo (Agilent Technologies) using the manufacturer’s instructions. PCR conditions were 95°Cfor 15 s, 74°C for 45 s and 68°C for 26
min. This cycle was repeated 25 times. Mutations were confirmed by sequencing done by Eton Bioscience.
1.5.3 In vitro transcription and RNA transfection
Infectious RNA was transcribed in vitro using SP6 RNA polymerase (New England Biolabs) following the manufacturer’s instructions. The transcripts were electroporated into
33 1.5.4 Amplification of Viral products with Infection
At least 1 mL of viral sample is required for the infection for a ~90% confluent cell monolayer in a T-75 cm2 flask. The amount of virus needed it dependent on the desired multiplicity of infection (MOI). U4.4 cells are starved of serum for one hour before the infection to allow the cells to better adhere to the bottom of the flask. Cells were rocked slowly at room temperature for an hour, allowing the virus to adsorb into the cells. Inoculum is subsequently removed and replaced with complete media. Virus is harvested 24 hours post-infection for BHK cells and 48 hours post-infection for U4.4 cells.
1.5.5 Virus Purification and particle/pfu determination
Viral samples were spun to equilibrium on a potassium tartrate step gradient. The gradient was formed by layering 15% potassium tartrate onto 35% potassium tartrate. The gradients were spun overnight at 24,000 rpm in a Beckman SW-28 rotor at 4°C. The viral band was collected before being layered onto a 15-35% linear potassium tartrate gradient. The samples were spun for 4 hours at 26,000 rpm in a Beckman SW-rotor at 4°C. Collected viral band was titered by plaque assay. The amount of viral protein was determined using Micro BCA protein assay reagent kit (Pierce), following the manufacturer’s instructions. Total number of particles
per infectious particle was determined by dividing the total number of particles by the number of infectious particles produced.
1.5.6 Virus Titration via Plaque Assay
35 REFERENCES
Akira, S., Uematsu, S., & Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell,
124(4), 783–801. https://doi.org/10.1016/j.cell.2006.02.015
Alcon, S., Talarmin, A., Debruyne, M., Falconar, A., Deubel, V., & Flamand, M. (2002). Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. Journal of Clinical Microbiology,
40(2), 376–381. https://doi.org/10.1128/JCM.40.2.376
Anthony, R. P., & Brown, D. T. (1991). Protein-protein interactions in an alphavirus membrane.
Journal of Virology, 65(3), 1187–1194. Retrieved from
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=239885&tool=pmcentrez&rend ertype=abstract
Bessaud, M., Peyrefitte, C. N., Pastorino, B. A. M., Tock, F., Merle, O., Colpart, J. J., …
Grandadam, M. (2006). Chikungunya virus strains, reunion island outbreak [2]. Emerging Infectious Diseases, 12(10), 1604–1606.
Brown, R. S., Wan, J. J., & Kielian, M. (2018). The alphavirus exit pathway: What we know and what we wish we knew. Viruses, 10(2). https://doi.org/10.3390/v10020089
Carleton, M., Lee, H., Mulvey, M., & Brown, D. T. (1997). Role of glycoprotein PE2 in formation and maturation of the Sindbis virus spike. J Virol, 71(2), 1558–1566. Retrieved from
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation &list_uids=8995682
36 Lipsitch, M. (2016). Systematic analysis of protein identity between Zika virus and other arthropod-borne viruses. Bulletin of the World Health Organization, 95(7), 517-525I. https://doi.org/10.2471/blt.16.182105
Chen, L., Wang, M., Zhu, D., Sun, Z., Ma, J., Wang, J., … Zhang, X. (2018). Implication for
alphavirus host-cell entry and assembly indicated by a 3.5Å resolution cryo-EM structure.
Nature Communications, 9(1). https://doi.org/10.1038/s41467-018-07704-x
Chen, M., Granger, A. J., VanBrocklin, M. W., Payne, W. S., Hunt, H., Zhang, H., … Holmen,
S. L. (2007). Inhibition of avian leukosis virus replication by vector-based RNA interference. Virology, 365(2), 464–472. https://doi.org/10.1016/j.virol.2007.04.013
Chen, M., Payne, W. S., Hunt, H., Zhang, H., Holmen, S. L., & Dodgson, J. B. (2008). Inhibition of Marek’s disease virus replication by retroviral vector-based RNA interference. Virology,
377(2), 265–272. https://doi.org/10.1016/j.virol.2008.03.019
Cohen, P. (2014). The TLR and IL-1 signalling network at a glance. Journal of Cell Science,
127(11), 2383–2390. https://doi.org/10.1242/jcs.149831
Counotte, M. J., Egli-Gany, D., Riesen, M., Abraha, M., Porgo, T. V., Wang, J., & Low, N. (2018). Zika virus infection as a cause of congenital brain abnormalities and Guillain-Barré syndrome: From systematic review to living systematic review. F1000Research, 7(0), 196. https://doi.org/10.12688/f1000research.13704.1
Dick, G., Kitchen, S., & Haddow, A. (1952). Zika virus. I. Isolations and serological specificity.
Transactions of The Royal Society of Tropical Medicine and Hygiene, 46(5), 509–520. Retrieved from https://doi.org/10.1016/0035-9203(52)90042-4
37 Virus from Recife, Brazil. PLoS Neglected Tropical Diseases, 10(10), 1–20.
https://doi.org/10.1371/journal.pntd.0005048
Eisen, L., & Moore, C. G. (2013). Aedes (Stegomyia) aegypti in the continental United States: a vector at the cool margin of its geographic range. Journal of Medical Entomology, 50(3), 467–478. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/23802440
Fernandez-Garcia, M. D., Mazzon, M., Jacobs, M., & Amara, A. (2009). Pathogenesis of Flavivirus Infections: Using and Abusing the Host Cell. Cell Host and Microbe, 5(4), 318– 328. https://doi.org/10.1016/j.chom.2009.04.001
Ferreira, D., Hernandez, R., Horton, M., & Brown, D. T. (2003). Morphological variants of sindbis virus produced by a mutation in the capsid protein. Virology, 307(1), 54–66. https://doi.org/10.1016/S0042-6822(02)00034-X
Ghose, J., Sinha, M., Das, E., Jana, N. R., & Bhattacharyya, N. P. (2011). Regulation of miR-146a by RelA/NFkB and p53 in STHdh q111/Hdh q111 cells, a cell model of Huntington’s
disease. PLoS ONE, 6(8). https://doi.org/10.1371/journal.pone.0023837
Ghosh, S., & Karin, M. (2002). Missing pieces in the NF-kappaB puzzle. Cell, 109 Suppl, S81-96. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11983155
Gong, Z., Xu, X., & Han, G. Z. (2017). The diversification of Zika virus: Are there two distinct lineages? Genome Biology and Evolution, 9(11), 2940–2945.
https://doi.org/10.1093/gbe/evx223
Guo, H.-Y., Zhang, X.-C., & Jia, R.-Y. (2018). Toll-Like Receptors and RIG-I-Like Receptors Play Important Roles in Resisting Flavivirus. Journal of Immunology Research, 2018, 1–7. https://doi.org/10.1155/2018/6106582
38 C. (2016). Genetic Characterization of Spondweni and Zika Viruses and Susceptibility of Geographically Distinct Strains of Aedes aegypti, Aedes albopictus and Culex
quinquefasciatus (Diptera: Culicidae) to Spondweni Virus. PLoS Neglected Tropical Diseases, 10(10), 1–13. https://doi.org/10.1371/journal.pntd.0005083
He, L., Meilleur, F., Piper, A., Heller, W. T., Hernandez, R., Brown, D. T., & Myles, D. A. A. (2010). The Structure of Sindbis Virus Produced from Vertebrate and Invertebrate Hosts as Determined by Small-Angle Neutron Scattering. Journal of Virology, 84(10), 5270–5276. https://doi.org/10.1128/jvi.00044-10
He, X., Jing, Z., & Cheng, G. (2014). MicroRNAs: New regulators of toll-like receptor signalling pathways. BioMed Research International, 2014.
https://doi.org/10.1155/2014/945169
Heinrich, P. C., Behrmann, I., Haan, S., Hermanns, H. M., & Uller-newen, G. M. (2003). Principles of IL 6 type cytokine signaling and its regulating. Biochem J, 374, 1–20. Retrieved from https://sci-hub.tw/10.1042/bj20030407
Hernandez, R., Ferreira, D., Sinodis, C., Litton, K., & Brown, D. T. (2005). Single Amino Acid Insertions at the Junction of the Sindbis Virus E2 Transmembrane Domain and Endodomain Disrupt Virus Envelopment and Alter Infectivity. Journal of Virology, 79(12), 7682–7697. https://doi.org/10.1128/jvi.79.12.7682-7697.2005
Hernandez, R., Lee, H., Nelson, C., & Brown, D. T. (2002). A Single Deletion in the Membrane-Proximal Region of the Sindbis Virus Glycoprotein E2 Endodomain Blocks Virus
Assembly. Journal of Virology, 74(9), 4220–4228. https://doi.org/10.1128/jvi.74.9.4220-4228.2000
39 in the Transmembrane Domain of a Sindbis Virus Glycoprotein Alter Virus Infectivity, Stability, and Host Range. Journal of Virology, 77(23), 12710–12719.
https://doi.org/10.1128/jvi.77.23.12710-12719.2003
Hsu, P. W. C., Lin, L. Z., Hsu, S. Da, Hsu, J. B. K., & Huang, H. Da. (2007). ViTa: Prediction of host microRNAs targets on viruses. Nucleic Acids Research, 35(SUPPL. 1), 381–385. https://doi.org/10.1093/nar/gkl1009
Hu, Y., & Sun, L. (2019). Systematic Analysis of Structure Similarity between Zika Virus and Other Flaviviruses. ACS Infectious Diseases. https://doi.org/10.1021/acsinfecdis.9b00047 Jana, A., Krett, N. L., Guzman, G., Khalid, A., Ozden, O., Staudacher, J. J., … Jung, B. (2017).
NFkB is essential for activin-induced colorectal cancer migration via upregulation of PI3K-MDM2 pathway. Oncotarget, 8(23), 37377–37393.
https://doi.org/10.18632/oncotarget.16343
Jopling, C., Yi, M., Lancaster, A., Lemon, S., & Sarnow, P. (2005). Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science, 309(September), 1577. Josseran, L., Paquet, C., Zehgnoun, A., Caillere, N., Le Tertre, A., Solet, J. L., & Ledrans, M.
(2006). Chikungunya disease outbreak, Reunion Island [1]. Emerging Infectious Diseases,
12(12), 1994–1995. https://doi.org/10.3201/eid1212.060710
K.M., O., R., L., W.J., E., P., M., A., K., R.M., E., … O.J., B. (2018). Projecting the end of the
Zika virus epidemic in Latin America: A modelling analysis. BMC Medicine, 16(1), 180. https://doi.org/http://dx.doi.org/10.1186/s12916-018-1158-8
Karin, M., & Lin, A. (2002). NF-kappa B at the crossroads of life and death. Nature Immunology, 3(3), 221–227.
40 Ross River Virus E1 That Allow Efficient Budding of Chimeric Viruses. Journal of
Virology, 74(6), 2663–2670. https://doi.org/10.1128/jvi.74.6.2663-2670.2000 Kishimoto, T. (2004). INTERLEUKIN-6: From Basic Science to Medicine—40 Years in
Immunology. Annual Review of Immunology, 23(1), 1–21. https://doi.org/10.1146/annurev.immunol.23.021704.115806
Kishimoto, T. (2010). IL-6: From its discovery to clinical applications. International Immunology, 22(5), 347–352. https://doi.org/10.1093/intimm/dxq030
La Linn, M., Gardner, J., Warrilow, D., Darnell, G. A., McMahon, C. R., Field, I., … Suhrbier, A. (2002). Arbovirus of Marine Mammals: a New Alphavirus Isolated from the Elephant Seal Louse, Lepidophthirus macrorhini. Journal of Virology, 75(9), 4103–4109.
https://doi.org/10.1128/jvi.75.9.4103-4109.2001
Lanciotti, R. S., & Lambert, A. J. (2016). Phylogenetic analysis of Chikungunya virus strains circulating in the Western Hemisphere. American Journal of Tropical Medicine and Hygiene, 94(4), 800–803. https://doi.org/10.4269/ajtmh.15-0375
Lanciotti, R. S., Lambert, A. J., Holodniy, M., Saavedra, S., & del Carmen Castillo Signor, L. (2016a). Countering Zika in Latin America: Epidemic dynamics are key and data gaps must be addressed. Science, 353(6297), 353–354. https://doi.org/10.1016/S2214-109X(16)30265-0.Cost-effectiveness
Lanciotti, R. S., Lambert, A. J., Holodniy, M., Saavedra, S., & del Carmen Castillo Signor, L. (2016b). Phylogeny of zika virus in western Hemisphere, 2015. Emerging Infectious Diseases, 22(5), 933–935. https://doi.org/10.3201/eid2205.160065
41 Liljeström, P., & Garoff, H. (1991). Internally located cleavable signal sequences direct the
formation of Semliki Forest virus membrane proteins from a polyprotein precursor. Journal of Virology, 65(1), 147–154. Retrieved from
http://www.ncbi.nlm.nih.gov/pubmed/1985194%0Ahttp://www.pubmedcentral.nih.gov/arti clerender.fcgi?artid=PMC240499
Lopez, S., Yao, J. S., Kuhn, R. J., Strauss, E. G., & Strauss, J. H. (1994).
Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. Journal of Virology, 68(3), 1316–1323. Retrieved from
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=236585&tool=pmcentrez&rend ertype=abstract%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/7508993%5Cnhttp://www.pub medcentral.nih.gov/articlerender.fcgi?artid=PMC236585
Lowe, R., Barcellos, C., Brasil, P., Cruz, O. G., Honório, N. A., Kuper, H., & Carvalho, M. S. (2018). The zika virus epidemic in brazil: From discovery to future implications.
International Journal of Environmental Research and Public Health, 15(1). https://doi.org/10.3390/ijerph15010096
Lwande, O., Obanda, V., Bucht, G., Mosomtai, G., Otieno, V., Ahlm, C., & Evander, M. (2015).
Global emergence of Alphaviruses that cause arthritis in humans. 1, 1–10.
Moehring, J. M., Inocencio, N. M., Robertson, B. J., & Moehring, T. J. (1993). Expression of mouse furin in a Chinese hamster cell resistant to Pseudomonas exotoxin A and viruses complements the genetic lesion. Journal of Biological Chemistry, 268(4), 2590–2594. MOUSSON, L., DAUGA, C., GARRIGUES, T., SCHAFFNER, F., VAZEILLE, M., &
42 variations . Genetical Research, 86(1), 1–11. https://doi.org/10.1017/s0016672305007627 Mukhopadhyay, S., Zhang, W., Gabler, S., Chipman, P. R., Strauss, E. G., Strauss, J. H., …
Rossmann, M. G. (2006). Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure, 14(1), 63–73.
https://doi.org/10.1016/j.str.2005.07.025
Mulvey, M., & Brown, D. T. (1996). Assembly of the Sindbis virus spike protein complex.
Virology, 219(1), 125–132. https://doi.org/10.1006/viro.1996.0229
Musso D, G. D. (2016). Zika virus. Clinical Microbiology Reviews, 30(2), 569–572. https://doi.org/10.1128/CMR.00072-15.Address
Nahid, M. A., Satoh, M., & Chan, E. K. L. (2015). Interleukin 1β-Responsive MicroRNA-146a Is Critical for the Cytokine-Induced Tolerance and Cross-Tolerance to Toll-Like Receptor Ligands. Journal of Innate Immunity, 7(4), 428–440. https://doi.org/10.1159/000371517 Oehler, E., Watrin, L., Larre, P., Lastère, S., Valour, F., Baudouin, L., … Musso, D. (2014). Zika
virus infection complicated by Guillain-Barré. Euro Surveillance, 1(December 2013), 7–9. Paredes, A. M., Brown, D. T., Rothnagel, R., Chiu, W., Schoepp, R. J., Johnston, R. E., &
Prasad, B. V. (1993). Three-dimensional structure of a membrane-containing virus.
Proceedings of the National Academy of Sciences, 90(19), 9095–9099. https://doi.org/10.1073/pnas.90.19.9095
Pletnev, S. V, Zhang, W., Mukhopadhyay, S., Fisher, B. R., Hernandez, R., Brown, D. T., …
https://ac.els-43 cdn.com/S0092867401003026/1-s2.0-S0092867401003026-main.pdf?_tid=01efc21b-bfb2-4a53-9113-d58f666fc31b&acdnat=1552282547_5e2e374fa83ddf6ce99bad648151f168 Ramsey, J., & Mukhopadhyay, S. (2017). Disentangling the frames, the state of research on the
alphavirus 6K and TF proteins. Viruses, 9(8), 1–21. https://doi.org/10.3390/v9080228 Rice, C. M., Bell, J. R., Hunkapiller, M. W., Strauss, E. G., & Strauss, J. H. (1982). Isolation and
characterization of the hydrophobic COOH-terminal domains of the Sindbis virion glycoproteins. Journal of Molecular Biology, 154(2), 355–378.
https://doi.org/10.1016/0022-2836(82)90069-9
Rice, C. M., Levis, R., Strauss, J. H., & Huang, H. V. (1987). Production of Infectious RNA Transcripts from Sindbis Virus cDNA Clones: Mapping of Lethal Mutations, Rescue of a Temperature- Sensitive Marker, and In Vitro Mutagenesis To Generate Defined Mutants.
Journal of Virology, 61(12), 3809–3819. Retrieved from
http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=3479621&retmo de=ref&cmd=prlinks%5Cnpapers3://publication/uuid/4601BFB8-566E-4001-B05A-FEDC898A10F9
Rusca, N., & Monticelli, S. (2011). MiR-146a in Immunity and Disease. Molecular Biology International, 2011, 1–7. https://doi.org/10.4061/2011/437301
Sato, S., Sanjo, H., Takeda, K., Ninomiya-Tsuji, J., Yamamoto, M., Kawai, T., … Akira, S. (2005). Essential function for the kinase TAK1 in innate and adaptive immune responses.
Nature Immunology, 6(11), 1087–1095. https://doi.org/10.1038/ni1255
Schimmack, G., Schorpp, K., Kutzner, K., Gehring, T., Brenke, J. K., Hadian, K., & Krappmann, D. (2017). YOD1/TRAF6 association balances p62-dependent IL-1 signaling to NF-κB.
44 Schmidt-Arras, D., & Rose-John, S. (2016). IL-6 pathway in the liver: From physiopathology to
therapy. Journal of Hepatology, 64(6), 1403–1415. https://doi.org/10.1016/j.jhep.2016.02.004
Schuler-faccini, L., Ribeiro, E. M., Feitosa, I. M. L., Horovitz, D. D. G., & Cavalcanti, D. P. (2016). Possible association between Zika virus infection and microcephaly—Brazil. 65(3), 59–62.
Shi, Y., & Gao, G. F. (2017). Structural Biology of the Zika Virus. Trends in Biochemical Sciences, 42(6), 443–456. https://doi.org/10.1016/j.tibs.2017.02.009
Shimakami, T., Yamane, D., Jangra, R. K., Kempf, B. J., Spaniel, C., Barton, D. J., & Lemon, S. M. (2012). Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex.
Proceedings of the National Academy of Sciences, 109(3), 941–946. https://doi.org/10.1073/pnas.1112263109
Sinodis, C., Ferreira, D., Hernandez, R., Horton, M., Brown, D. T., & Yang, C. (2003). Deletions in the Transmembrane Domain of a Sindbis Virus Glycoprotein Alter Virus Infectivity, Stability, and Host Range. Journal of Virology, 77(23), 12710–12719.
https://doi.org/10.1128/jvi.77.23.12710-12719.2003
Sjoberg, M., & Garoff, H. (2003). Interactions between the Transmembrane Segments of the Alphavirus E1 and E2 Proteins Play a Role in Virus Budding and Fusion Mathilda. 77(6), 55–78. https://doi.org/10.1128/JVI.77.6.3441
Slon Campos, J. L., Mongkolsapaya, J., & Screaton, G. R. (2018). The immune response against flaviviruses. Nature Immunology, 19(11), 1189–1198. https://doi.org/10.1038/s41590-018-0210-3
45 Hydrophobic Anchors of Glycoproteins E2 and E1 Interact during Assembly of
Alphaviruses. Journal of Virology, 76(20), 10188–10194. https://doi.org/10.1128/jvi.76.20.10188-10194.2002
Strauss, J. H., & Strauss, E. G. (1994). The alphaviruses: gene expression, replication, and evolution. Microbiological Reviews, 58(3), 491–562. Retrieved from
http://www.ncbi.nlm.nih.gov/pubmed/7968923%0Ahttp://www.pubmedcentral.nih.gov/arti clerender.fcgi?artid=PMC372977
Suthar, M. S., Aguirre, S., & Fernandez-Sesma, A. (2013). Innate Immune Sensing of Flaviviruses. PLoS Pathogens, 9(9), e1003541.
https://doi.org/10.1371/journal.ppat.1003541
Taganov, K. D., Boldin, M. P., & Baltimore, D. (2007). MicroRNAs and Immunity: Tiny Players in a Big Field. Immunity, 26(2), 133–137. https://doi.org/10.1016/j.immuni.2007.02.005 Taylor, R. M., Hurlbut, H. S., Work, T. H., & Government, E. (1953). Sindbis virus: a newly
recognized arthropod-transmitted virus1. 844–862.
Temperley, N. D., Berlin, S., Paton, I. R., Griffin, D. K., & Burt, D. W. (2008). Evolution of the chicken Toll-like receptor gene family: A story of gene gain and gene loss. BMC Genomics,
9, 1–12. https://doi.org/10.1186/1471-2164-9-62
Trobaugh, D. W., Gardner, C. L., Sun, C., Haddow, A. D., Wang, E., Chapnik, E., … Klimstra,
W. B. (2014). RNA viruses can hijack vertebrate microRNAs to suppress innate immunity.
Nature, 506(7487), 245–248. https://doi.org/10.1038/nature12869
Trobaugh, D. W., & Klimstra, W. B. (2017). MicroRNA Regulation of RNA Virus Replication and Pathogenesis. 40(4), 1291–1296.
46 Valadão, A. L. C., Aguiar, R. S., & de Arruda, L. B. (2016). Interplay between inflammation and
cellular stress triggered by Flaviviridae viruses. Frontiers in Microbiology, 7(AUG), 1–19. https://doi.org/10.3389/fmicb.2016.01233
Virtue, A., Wang, H., & Yang, X. F. (2012). MicroRNAs and Toll-like receptor/interleukin-1 receptor signaling. Journal of Hematology and Oncology, 5(1), 1.
https://doi.org/10.1186/1756-8722-5-66
Virus, Z. (2017). Situation report Zika virus march 10. (MARCH), 1–5.
Volk, S. M., Chen, R., Tsetsarkin, K. A., Adams, A. P., Garcia, T. I., Sall, A. A., … Weaver, S.
C. (2010). Genome-Scale Phylogenetic Analyses of Chikungunya Virus Reveal
Independent Emergences of Recent Epidemics and Various Evolutionary Rates. Journal of Virology, 84(13), 6497–6504. https://doi.org/10.1128/jvi.01603-09
Whitehurst, C. B., Willis, J. H., Sinodis, C. N., Hernandez, R., & Brown, D. T. (2006). Single and multiple deletions in the transmembrane domain of the Sindbis virus E2 glycoprotein identify a region critical for normal virus growth. Virology, 347(1), 199–207.
https://doi.org/10.1016/j.virol.2005.11.029
WHO. (2016). RAPID RISK ASSESSMENT Zika virus disease epidemic : potential association
with microcephaly and Guillain-Barré syndrome ( 1 st update ) Main conclusions. European Centre for Disease Prevention and Control, (January), 1–20.
Wu, S., He, L., Li, Y., Wang, T., Feng, L., Jiang, L., … Huang, X. (2013). MiR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. Journal of Infection, 67(4), 329–341. https://doi.org/10.1016/j.jinf.2013.05.003
47 genetically divergent strains of Zika. Genome Announcements, 4(4), 1–4.
48
Title: The alphaviruses: gene expression, replication, and evolution.
Author: J H Strauss,E G Strauss Publication: Microbiology and Molecular
Biology Reviews Publisher: American Society for
Microbiology Date: Sep 1, 1994
Copyright © 1994, American Society for Microbiology
LOGIN
If you're a copyright.com user, you can login to RightsLink using your copyright.com credentials. Already a RightsLink user or want to learn more?
Permissions Request
ASM authorizes an advanced degree candidate to republish the requested material in his/her doctoral thesis or dissertation. If your thesis, or dissertation, is to be published commercially, then you must reapply for permission.
49
Title: Single and multiple deletions in the transmembrane domain of the Sindbis virus E2 glycoprotein identify a region critical for normal virus growth
Author: Christopher B. Whitehurst,John H. Willis,Christine N.
Sinodis,Raquel
Hernandez,Dennis T. Brown Publication: Virology
Publisher: Elsevier Date: 30 March 2006
Copyright © 2005 Elsevier Inc. All rights reserved.
Logged in as: Sophia Yang North Carolina State University
Order Completed
Thank you for your order.
This Agreement between North Carolina State University -- Sophia Yang ("You") and Elsevier ("Elsevier") consists of your license details and the terms and conditions provided by Elsevier and Copyright
Clearance Center.
Your confirmation email will contain your order number for future reference.
printable details
License Number 4596560391540
License date May 26, 2019
Licensed Content
Publisher Elsevier
Licensed Content
Publication Virology
Licensed Content Title Single and multiple deletions in the transmembrane domain of the Sindbis virus E2 glycoprotein
identify a region critical for normal virus growth
Licensed Content Author Christopher B. Whitehurst,John H. Willis,Christine N. Sinodis,Raquel Hernandez,Dennis T. Brown
Licensed Content Date Mar 30, 2006
Licensed Content Volume 347
Licensed Content Issue 1
Licensed Content Pages 9
Type of Use reuse in a thesis/dissertation
Portion figures/tables/illustrations
Number of
figures/tables/illustrations 3
Format both print and electronic
Are you the author of this
Elsevier article? No
Will you be translating? No
50
Title of your
thesis/dissertation Arbovirus structure and interaction with host cells
Expected completion date Jun 2019
Estimated size (number
of pages) 80
Requestor Location North Carolina State University
120 Broughton Drive 128 Polk Hall
RALEIGH, NC 27695 United States Attn: Sophia Yang
Publisher Tax ID 98-0397604
51
Title: Deletions in the Transmembrane Domain of a Sindbis Virus Glycoprotein Alter Virus Infectivity, Stability, and Host Range
Author: Raquel Hernandez,Christine Sinodis,Michelle Horton,Davis Ferreira,Chunning Yang,Dennis T. Brown
Publication: Journal of Virology Publisher: American Society for
Microbiology Date: Nov 10, 2003
Copyright © 2003, American Society for Microbiology
Logged in as: Sophia Yang North Carolina State University
Permissions Request
ASM authorizes an advanced degree candidate to republish the requested material in his/her doctoral thesis or dissertation. If your thesis, or dissertation, is to be published commercially, then you must reapply for permission.
52 CHAPTER 2
Role of miR-146a on Zika Virus Pathophysiology
Sophia Yang, Davis Ferreira, Julie A. Hicks, Hsiao-Ching Liu, Dennis T. Brownand Raquel Hernandez
53 2.1 Abstract
Zika virus (ZIKV) is a Flavivirus with an RNA dense core and is transmitted to humans mainly by Aedes aegypti mosquitos. It was first isolated in the Zika forest of Uganda, Africa in 1947 (Dick et al., 1952), and was found to produce mild symptoms including fever, headache, malaise, rash and conjunctivitis. As the virus expanded its range 50-100 years ago, it migrated from East Africa into Asia, evolving into a unique Asian genotype (Lanciotti & Lambert, 2016; Lanciotti et al., 2016b; Volk et al., 2010). This evolution was accompanied with increased virus pathogenicity, as the disease progressed from a mild to non-perceptible infection to a highly pathogenic disease producing severe encephalitic symptoms. This evolved virus also expanded its mode of infection from strictly mosquito vectored to acquiring the ability for direct person-to-person, sexual and vertical transmission from mother to fetus. The genomic sequence analysis of non- epidemic (African, AF) and epidemic strains (Asian, AS) are similar, demonstrating
54 regulate microRNA expression and thus leading to a difference within host cell immune
response.
Single stranded RNA viruses have been shown to use microRNA (miRNA) to alter host cell immune response to viral infection (Jopling et al., 2005; Trobaugh et al., 2014). To
investigate how the virus advanced from non-pathogenic to pathogenic, the differences in profile of the miR-146a were investigated. miRNA146a was chosen as a likely target because it is known to regulate the toll-like receptor innate immune response (Virtue et al., 2012). This pathway is altered by flavivirus infections of West Nile virus (WNV), Japanese encephalitis virus (JEV) dengue (DENV) and ZIKV (Guo et al., 2018). Human monocytes and macrophages, MФ, were infected with the AF (MR766) and AS (PRVABC59) Zika strains. Using miR-146a
as our target regulator, the expression of select genes associated with miRNA146a expression during infections of human THP-1 cells with an AF or AS strains was studied. It was found that there are profound differences in the expression of ZIKV, miR-146a regulated genes between the two stains. Several of the genes are found to be associated in pathways leading to host cell response to infection. Such pathways include those resulting in inflammation, innate and
55 2.2 Introduction
Zika virus (ZIKV) is a Flavivirus with a positive sense, single stranded RNA genome. Other medically important viruses within in this family include dengue (DENV), yellow fever virus (YF) and Japanese encephalitis virus (JEV). Flaviviruses are known to cause hemorrhagic, arthritic and encephalitic disease in humans. The virus is mainly spread by the Aedes aegypti
mosquito which originated in Africa (Mousson et al., 2005), but is now found in tropical, subtropical and temperate regions around the world (Eisen & Moore, 2013).
There are two lineages of the ZIKV, African (AF) and Asian (AS) (Gong, Xu, & Han, 2017) which share 96% sequence identity (Lanciotti et al., 2016b). MR766 is an African strain of ZIKV that was first identified in Uganda in 1947 (Dick et al., 1952). This strain is the ZIKV reference strain and it has close sequence fidelity to the Spondweni root virus (Figure 2.1) (Haddow et al., 2016). MR766 is non-pathogenic and produces sub-clinical symptoms in the infected host. The first AS strain was identified in Malaysia in 1966 (Lee & Ng, 2018) and designated to a separated ZIKV clade. Early ZIKV strains circulating throughout Africa into Asia were not pathogenic, producing sub-clinical symptoms. This lack of clinical history is documented by the presence of ZIKV seropositive individuals found in flavivirus endemic regions. The virus evolved, during its transmittal to Asia, to become a potent pathogen,