organic papers
Acta Cryst.(2007). E63, o1867–o1868 doi:10.1107/S1600536807011683 Nget al. C
13H9BrOS
o1867
Acta Crystallographica Section EStructure Reports Online
ISSN 1600-5368
(2
E
)-1-(3-Bromophenyl)-3-(2-thienyl)prop-2-en-1-one
Shea-Lin Ng,aP. S. Patil,b
Ibrahim Abdul Razak,a
Hoong-Kun Fun,a*
H. B. Ramesh Babuband
S. M. Dharmaprakashb
aX-ray Crystallography Unit, School of Physics,
Universiti Sains Malaysia, 11800 USM, Penang, Malaysia, andbDepartment of Studies in Physics, Mangalore University,
Mangalagangotri, Mangalore 574 199, India.
Correspondence e-mail: hkfun@usm.my
Key indicators
Single-crystal X-ray study T= 100 K
Mean(C–C) = 0.002 A˚ Disorder in main residue Rfactor = 0.023 wRfactor = 0.070
Data-to-parameter ratio = 24.1
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
Received 9 March 2007 Accepted 13 March 2007
#2007 International Union of Crystallography All rights reserved
The thiophene ring of the title compound, C13H9BrOS, is
disordered. The disorder corresponds to an approximate 180
rotation of the thiophene ring with respect to the C—C bond linking it to the rest of the molecule. The crystal packing is stabilized by intermolecular C—H Br and C—H inter-actions, together with short S Br contacts.
Comment
Many chalcone derivatives crystallize as non-centrosymmetric structures and display significant non-linear optical (NLO) properties (Patil, Tehet al., 2006; Patil, Dharmaprakashet al., 2006; Shettigaret al., 2006; Patil, Nget al., 2007; Patil, Rosliet al., 2007). The single-crystal X-ray structural study of the title compound, (I), was undertaken in order to establish the structure and conformation of the various groups. Crystal-lization of the title compound in a centrosymmetric space group precludes second-order non-linear optical properties.
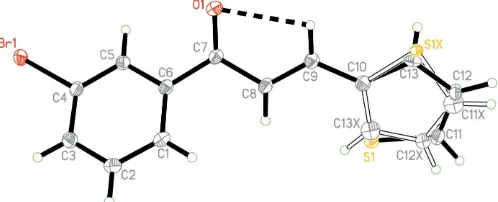
In (I) (Fig. 1), the thiophene ring is disordered over two sites (atoms of the minor occupancy component are labelled with the suffix X). The disorder corresponds to an approx-imate 180 rotation of the thiophene ring with respect to the
C9—C10 bond. Bond lengths and angles in (I) are comparable to those reported in a related structure (Nget al., 2006) and show normal values (Allen et al., 1987). The molecule is twisted with dihedral angles of 48.2 (2) and 47.1 (8),
[image:1.610.208.457.603.704.2]respec-tively, between the benzene ring and the thiophene rings C10– C13/S1 and C10/C11—C13X/S1X.
Figure 1
In the crystal packing, for the major component, there are no intermolecular interactions. However, for the minor component, molecules are linked by intermolecular C11X— H11B Br1iinteractions (Table 1) and S1X Br1(x,y,1 +z) short contacts [3.590 (9) A˚ ], forming sheet-like structures parallel to theacplane. The sheets are stacked along thebaxis (Fig. 2). In addition, the crystal packing is further stabilized by weak intermolecular C—H interactions involving the C1– C6 (centroidCg3), C10/S1/C11–C13 (centroidCg1) and C10/ S1X/C11X–C13X(centroidCg2) rings.
Experimental
2-Thiophenecarbaldehyde (0.1 mol) and 3-bromoacetophenone (0.1 mol) were strirred in ethanol (100 ml) at 298 K. 10 g of a 10% aqueous NaOH solution was added and the mixture was stirred for 2 h. The resulting precipitate was filtered off, washed with water and dried. The resulting crude product was recrystallized twice from acetone. The purity of the compound was checked by thin-layer chromatography. Crystals suitable for single-crystal X-ray diffraction experiments were grown by slow evaporation of an acetone solution.
Crystal data
C13H9BrOS
Mr= 293.17 Triclinic,P1 a= 5.8468 (1) A˚ b= 7.4089 (2) A˚ c= 13.1979 (3) A˚
= 100.215 (1)
= 91.086 (1)
= 90.174 (1)
V= 562.54 (2) A˚3 Z= 2
MoKradiation
= 3.81 mm1
T= 100.0 (1) K 0.460.340.10 mm
Data collection
Bruker SMART APEX2 CCD area-detector diffractometer Absorption correction: multi-scan
(SADABS; Bruker, 2005) Tmin= 0.296,Tmax= 0.702
17165 measured reflections 4018 independent reflections 3615 reflections withI> 2(I) Rint= 0.025
Refinement
R[F2> 2(F2)] = 0.023 wR(F2) = 0.070
S= 1.13 4018 reflections
167 parameters
H-atom parameters constrained
max= 0.72 e A˚
3
min=0.39 e A˚
[image:2.610.45.294.70.150.2]3
Table 1
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
C9—H9A O1 0.93 2.47 2.805 (2) 101 C11X—H11B Br1i
0.93 2.83 3.486 (19) 128 C2—H2A Cg1ii
0.93 2.90 3.544 (2) 128 C2—H2A Cg2ii
0.93 2.88 3.536 (9) 128 C5—H5A Cg1iii 0.93 2.85 3.561 (2) 134 C5—H5A Cg2iii
0.93 2.85 3.553 (8) 133 C9—H9A Cg3iv
0.93 3.07 3.633 (2) 121 C11—H11A Cg3v
0.93 3.04 3.581 (2) 118 C12X—H12B Cg3v
0.93 2.88 3.535 (16) 129
Symmetry codes: (i)x1;y;z1; (ii)xþ1;yþ1;z; (iii)xþ2;yþ2;z; (iv)xþ2;yþ1;z; (v)xþ1;yþ2;z.Cg1,Cg2 andCg3 are the centroids of the C10/S1/C11–C13, C10/S1X/C11X–C13Xand C1–C6 rings, respectively.
H atoms were placed in calculated positions and constrained to ride on their carrier atoms, with C—H = 0.93 A˚ and Uiso(H) = 1.2Ueq(C). The ratio of the refined site occupancies for the major and minor components of the disordered thiophene ring is 0.858 (3):0.142 (3); the C atoms of the minor disorder component were refined isotropically. Similarity and rigid-bond restraints were applied to the displacement parameters of the disordered atoms, and there were also geometrical restraints.
Data collection:APEX2(Bruker, 2005); cell refinement:APEX2; data reduction: SAINT (Bruker, 2005); program(s) used to solve structure: SHELXTL (Sheldrick, 1998); program(s) used to refine structure:SHELXTL; molecular graphics:SHELXTL; software used to prepare material for publication:SHELXTL,PARST(Nardelli, 1995) andPLATON(Spek, 2003).
The authors thank the Malaysian Government and Universiti Sains Malaysia for the Scientific Advancement Grant Allocation (SAGA) grant No. 304/PFIZIK/653003/ A118 and the Fundamental Research Grant Scheme (FRGS): 203/PFIZIK/671064. PSP thanks DRDO, Goverment of India, for a Junior Research Fellowship (JRF).
References
Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987).J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
Bruker (2005). APEX2 (Version 1.27), SAINT (Version V7.12A) and SADABS(Version 2004/1). Bruker AXS Inc., Madison, Wisconsin, USA. Nardelli, M. (1995).J. Appl. Cryst.28, 659.
Ng, S.-L., Razak, I. A., Fun, H.-K., Patil, P. S. & Dharmaprakash, S. M. (2006). Acta Cryst.E62, o4653–o4655.
Patil, P. S., Dharmaprakash, S. M., Fun, H.-K. & Karthikeyan, M. S. (2006).J. Cryst. Growth,297, 111–116.
Patil, P. S., Ng, S.-L., Razak, I. A., Fun, H.-K. & Dharmaprakash, S. M. (2007). Acta Cryst.E63, o59–o60.
Patil, P. S., Rosli, M. M., Fun, H.-K., Razak, I. A. & Dharmaprakash, S. M. (2007).Acta Cryst.E63, o785–o786.
Patil, P. S., Teh, J. B.-J., Fun, H.-K., Razak, I. A. & Dharmaprakash, S. M. (2006).Acta Cryst.E62, o3096–o3098.
Sheldrick, G. M. (1998).SHELXTL. Version 5.1. Bruker AXS Inc., Madison, Wisconsin, USA.
Shettigar, V., Patil, P. S., Naveen, S., Dharmaprakash, S. M., Sridhar, M. A. & Shashidra Prasad, J. (2006).J. Cryst. Growth,295, 44–49.
Spek, A. L. (2003).J. Appl. Cryst.36, 7–13. Figure 2
supporting information
sup-1 Acta Cryst. (2007). E63, o1867–o1868
supporting information
Acta Cryst. (2007). E63, o1867–o1868 [https://doi.org/10.1107/S1600536807011683]
(2
E
)-1-(3-Bromophenyl)-3-(2-thienyl)prop-2-en-1-one
Shea-Lin Ng, P. S. Patil, Ibrahim Abdul Razak, Hoong-Kun Fun, H. B. Ramesh Babu and S. M.
Dharmaprakash
(2E)-1-(3-bromophenyl)-3-(2-thienyl)prop-2-en-1-one
Crystal data
C13H9BrOS Mr = 293.17
Triclinic, P1 Hall symbol: -P 1
a = 5.8468 (1) Å
b = 7.4089 (2) Å
c = 13.1979 (3) Å
α = 100.215 (1)°
β = 91.086 (1)°
γ = 90.174 (1)°
V = 562.54 (2) Å3
Z = 2
F(000) = 292
Dx = 1.731 Mg m−3
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 9217 reflections
θ = 2.8–32.5°
µ = 3.81 mm−1 T = 100 K Block, yellow
0.46 × 0.34 × 0.10 mm
Data collection
Bruker SMART APEX2 CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
Detector resolution: 8.33 pixels mm-1 ω scans
Absorption correction: multi-scan (SADABS; Bruker, 2005)
Tmin = 0.296, Tmax = 0.702
17165 measured reflections 4018 independent reflections 3615 reflections with I > 2σ(I)
Rint = 0.025
θmax = 32.5°, θmin = 2.8° h = −8→8
k = −11→11
l = −19→19
Refinement
Refinement on F2
Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.023 wR(F2) = 0.070 S = 1.13 4018 reflections 167 parameters 107 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained
w = 1/[σ2(F
o2) + (0.0319P)2 + 0.3942P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.72 e Å−3
Special details
Experimental. The data was collected with the Oxford Cyrosystem Cobra low-temperature attachment.
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full
covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq Occ. (<1)
Br1 1.17143 (3) 0.72219 (2) 0.451605 (12) 0.01934 (5) O1 1.1990 (2) 0.71774 (18) 0.03823 (9) 0.0203 (2) C1 0.6886 (3) 0.6111 (2) 0.16180 (13) 0.0168 (3)
H1A 0.5901 0.5912 0.1047 0.020*
C2 0.6196 (3) 0.5659 (2) 0.25453 (13) 0.0178 (3)
H2A 0.4764 0.5130 0.2586 0.021*
C3 0.7627 (3) 0.5990 (2) 0.34081 (13) 0.0177 (3)
H3A 0.7164 0.5698 0.4030 0.021*
C4 0.9767 (3) 0.6768 (2) 0.33263 (12) 0.0155 (3) C5 1.0520 (3) 0.7199 (2) 0.24070 (12) 0.0152 (3)
H5A 1.1968 0.7701 0.2366 0.018*
C6 0.9056 (3) 0.6862 (2) 0.15436 (12) 0.0147 (3) C7 0.9931 (3) 0.7240 (2) 0.05417 (12) 0.0152 (3) C8 0.8252 (3) 0.7678 (2) −0.02296 (12) 0.0166 (3) H8A 0.6739 0.7920 −0.0049 0.020* C9 0.8921 (3) 0.7726 (2) −0.11926 (12) 0.0161 (3) H9A 1.0432 0.7415 −0.1344 0.019* C10 0.7520 (3) 0.8212 (2) −0.20139 (12) 0.0157 (3)
supporting information
sup-3 Acta Cryst. (2007). E63, o1867–o1868
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
Br1 0.01880 (9) 0.02695 (9) 0.01295 (7) −0.00228 (6) −0.00175 (5) 0.00569 (5) O1 0.0144 (5) 0.0291 (6) 0.0178 (5) 0.0011 (4) −0.0008 (4) 0.0051 (4) C1 0.0141 (7) 0.0169 (6) 0.0188 (7) 0.0007 (5) −0.0024 (5) 0.0019 (5) C2 0.0142 (7) 0.0160 (6) 0.0231 (7) 0.0001 (5) 0.0014 (5) 0.0034 (5) C3 0.0177 (7) 0.0179 (7) 0.0181 (7) 0.0019 (5) 0.0014 (5) 0.0048 (5) C4 0.0153 (7) 0.0175 (6) 0.0138 (6) 0.0025 (5) −0.0012 (5) 0.0029 (5) C5 0.0152 (7) 0.0154 (6) 0.0149 (6) 0.0011 (5) 0.0009 (5) 0.0028 (5) C6 0.0148 (7) 0.0141 (6) 0.0149 (6) 0.0020 (5) −0.0010 (5) 0.0019 (5) C7 0.0148 (7) 0.0159 (6) 0.0144 (6) 0.0008 (5) −0.0020 (5) 0.0018 (5) C8 0.0156 (7) 0.0186 (7) 0.0151 (6) 0.0034 (5) −0.0025 (5) 0.0022 (5) C9 0.0158 (7) 0.0161 (6) 0.0164 (6) 0.0013 (5) −0.0009 (5) 0.0035 (5) C10 0.0158 (7) 0.0151 (6) 0.0165 (6) 0.0012 (5) −0.0001 (5) 0.0033 (5) S1 0.0142 (3) 0.0188 (3) 0.0159 (2) 0.0030 (2) 0.00010 (18) 0.00280 (16) C11 0.0183 (10) 0.0190 (9) 0.0156 (10) −0.0008 (6) −0.0033 (8) 0.0047 (8) C12 0.0191 (12) 0.0207 (11) 0.0115 (9) −0.0016 (10) −0.0018 (8) 0.0038 (7) C13 0.010 (2) 0.0156 (14) 0.0203 (17) 0.0011 (14) 0.0046 (12) 0.0013 (11) S1X 0.009 (3) 0.022 (2) 0.011 (2) 0.007 (2) 0.0016 (18) 0.0037 (15)
Geometric parameters (Å, º)
Br1—C4 1.9022 (15) C9—H9A 0.9300
O1—C7 1.225 (2) C10—C13 1.394 (6)
C1—C2 1.391 (2) C10—C13X 1.407 (15)
C1—C6 1.396 (2) C10—S1X 1.608 (8)
C1—H1A 0.9300 C10—S1 1.7158 (18)
C2—C3 1.386 (2) S1—C11 1.714 (3)
C2—H2A 0.9300 C11—C12 1.364 (4)
C3—C4 1.390 (2) C11—H11A 0.9300
C3—H3A 0.9300 C12—C13 1.398 (7)
C4—C5 1.387 (2) C12—H12A 0.9300
C5—C6 1.398 (2) C13—H13A 0.9300
C5—H5A 0.9300 S1X—C11X 1.749 (15)
C6—C7 1.498 (2) C11X—C12X 1.345 (15)
C7—C8 1.479 (2) C11X—H11B 0.9300
C8—C9 1.343 (2) C12X—C13X 1.395 (17)
C8—H8A 0.9300 C12X—H12B 0.9300
C9—C10 1.443 (2) C13X—H13B 0.9300
C2—C1—C6 120.07 (15) C13—C10—C9 127.0 (3)
C2—C1—H1A 120.0 C13X—C10—C9 124.0 (8)
C6—C1—H1A 120.0 C13X—C10—S1X 116.1 (8) C3—C2—C1 120.50 (15) C9—C10—S1X 119.8 (3)
C3—C2—H2A 119.8 C13—C10—S1 109.3 (3)
C1—C2—H2A 119.8 C9—C10—S1 123.70 (12)
C2—C3—H3A 120.6 C11—S1—C10 92.44 (10)
C4—C3—H3A 120.6 C12—C11—S1 112.30 (19)
C5—C4—C3 122.07 (15) C12—C11—H11A 123.8 C5—C4—Br1 119.54 (12) S1—C11—H11A 123.8 C3—C4—Br1 118.39 (12) C11—C12—C13 111.5 (3) C4—C5—C6 118.57 (15) C11—C12—H12A 124.2
C4—C5—H5A 120.7 C13—C12—H12A 124.2
C6—C5—H5A 120.7 C10—C13—C12 114.5 (4)
C1—C6—C5 120.03 (14) C10—C13—H13A 122.8 C1—C6—C7 121.77 (14) C12—C13—H13A 122.8 C5—C6—C7 118.13 (14) C10—S1X—C11X 89.5 (7) O1—C7—C8 122.02 (15) C12X—C11X—S1X 112.9 (13) O1—C7—C6 119.78 (14) C12X—C11X—H11B 123.5 C8—C7—C6 118.20 (14) S1X—C11X—H11B 123.5 C9—C8—C7 119.71 (15) C11X—C12X—C13X 111.6 (14)
C9—C8—H8A 120.1 C11X—C12X—H12B 124.2
C7—C8—H8A 120.1 C13X—C12X—H12B 124.2
C8—C9—C10 126.24 (15) C12X—C13X—C10 109.6 (13)
C8—C9—H9A 116.9 C12X—C13X—H13B 125.2
C10—C9—H9A 116.9 C10—C13X—H13B 125.2
C13—C10—C13X 109.0 (9)
C6—C1—C2—C3 1.6 (2) C13—C10—S1—C11 0.5 (4) C1—C2—C3—C4 −0.5 (2) C9—C10—S1—C11 −179.58 (15) C2—C3—C4—C5 −0.8 (2) S1X—C10—S1—C11 2.3 (5) C2—C3—C4—Br1 179.88 (12) C10—S1—C11—C12 −0.2 (2) C3—C4—C5—C6 1.0 (2) S1—C11—C12—C13 −0.1 (5) Br1—C4—C5—C6 −179.69 (11) C13X—C10—C13—C12 −1.2 (14) C2—C1—C6—C5 −1.4 (2) C9—C10—C13—C12 179.4 (3) C2—C1—C6—C7 175.45 (14) S1X—C10—C13—C12 −168 (7) C4—C5—C6—C1 0.1 (2) S1—C10—C13—C12 −0.6 (6) C4—C5—C6—C7 −176.85 (13) C11—C12—C13—C10 0.5 (7) C1—C6—C7—O1 −150.63 (15) C13—C10—S1X—C11X 10 (6) C5—C6—C7—O1 26.3 (2) C13X—C10—S1X—C11X −3.9 (17) C1—C6—C7—C8 29.3 (2) C9—C10—S1X—C11X 178.4 (10) C5—C6—C7—C8 −153.80 (14) S1—C10—S1X—C11X −3.4 (12) O1—C7—C8—C9 10.9 (2) C10—S1X—C11X—C12X 3 (2) C6—C7—C8—C9 −169.04 (14) S1X—C11X—C12X—C13X −1 (3) C7—C8—C9—C10 −176.94 (15) C11X—C12X—C13X—C10 −2 (3) C8—C9—C10—C13 −168.2 (4) C13—C10—C13X—C12X 2 (2) C8—C9—C10—C13X 12.5 (14) C9—C10—C13X—C12X −178.5 (11) C8—C9—C10—S1X −170.1 (6) S1X—C10—C13X—C12X 4 (2) C8—C9—C10—S1 11.8 (2)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
supporting information
sup-5 Acta Cryst. (2007). E63, o1867–o1868
C11X—H11B···Br1i 0.93 2.83 3.486 (19) 128
C2—H2A···Cg1ii 0.93 2.90 3.544 (2) 128
C2—H2A···Cg2ii 0.93 2.88 3.536 (9) 128
C5—H5A···Cg1iii 0.93 2.85 3.561 (2) 134
C5—H5A···Cg2iii 0.93 2.85 3.553 (8) 133
C9—H9A···Cg3iv 0.93 3.07 3.633 (2) 121
C11—H11A···Cg3v 0.93 3.04 3.581 (2) 118
C12X—H12B···Cg3v 0.93 2.88 3.535 (16) 129