JOURNALOFVIROLOGY, Aug. 1979, p. 265-276
0022-538X/79/08.0265/12$02.00/0 Vol. 31, No. 2
Physical Mapping of
paar
Mutations of Herpes Simplex Virus
Type
1
and
Type
2
by Intertypic
Marker Rescue
PIERRE CHARTRAND, NIGEL D. STOW, MORAG C. TIMBURY, ANDNEIL M. WILKIE* Medical ResearchCouncilVirology Unit and Department of Virology, University of Glasgow, Glasgow Gll
5JR,
Scotlaud
Received forpublication15January 1979
Mutations (paar) in herpes simplex virus (HSV) which confer resistance to
phosphonoacetic acid involve genes associated with virus-induced DNA
poly-merase activity. Two mutantsof HSV (HSV-1 tsH and HSV-2 ts6) produce a
thermolabile DNA polymerase activity. In this study, the tslesions presentin these mutants and those present in two independent phosphonoacetic acid-resistantmutantsof HSV-1 andHSV-2 (paar_1 andpaar-2) have been physically mapped byrestriction endonucleaseanalysis of recombinants produced between HSV-1 and HSV-2 by intertypic markerrescue.Allfourmutations mapped within
a 3.3-kilobase pair region around map unit40. The accuracy of the method is
reflected by themapping results for tsH andpaar-2, whichwerefoundtoliein thesame1.3-kilobase pair region.paad-1wasfoundtolietothe right ofts6.
Virus-induced DNA polymerase is thought to have a molecular weight of 150,000, necessitating a gene with a coding capacity of 4.6 kilobase pairs. The four
mutationsmappedinthisstudyall liewithinaregionsmallerthanthis, but the
results donotyetprovethatallfour lesions residein thisor anysinglegene.
Phosphonoacetic acid (PAA) inhibits herpes
simplex virus (HSV) replicationin vivo(17),and Maoetal. (12) showed that thedrugblocksthe activity of virus-induced DNA polymerase in vitro. Mutantsof HSV-1 and HSV-2whichare resistant tothe drug (PAAr) have been isolated in several laboratories (6-8), and studies using these mutantshave indicated a direct
involve-mentofvirus-induced DNApolymerase activity
inthemechanismof inhibition.
Hay andSubak-Sharpe (6) firstshowedthat the DNApolymeraseactivity induced by PAA'
mutants was moreresistant to PAA invitrothan that ofwild-type HSV.Resistancetoincreasing concentrationsof thedrugcanbeacquiredby a
multiple-stepprocess (implying several consec-utivemutations), and the increased level of re-sistance in vivoisreflectedin theresistance of DNApolymerase activity invitro (7). Genetic analysis of these consecutive mutations indi-cated very close linkage on the viral genome. Joffre et al. (8) also reported the inability to segregatethe lesions of threeindependently iso-latedPAArmutants. The mostdirect evidence thatthePAAresistancemutation
(paar)
occurs inthestructuralgene of thevirus-inducedDNA polymerase comesfrom the analysis of HSV-1 tsD9 (8, 20, 21). Thistemperature-sensitive (ts) mutant is resistanttoPAA,although it was notselectedfor thatphenotype, andrevertantsfor the tsphenotypealsorevert toPAAsensitivity
(PAA5).
TheDNApolymeraseactivityinducedby HSV-1 tsD9 is thermolabile in vivo (1, 8). Recently, Purifoy et al. (20) have shown that
extensively purifiedDNA
polymerase
activityofHSV-1 tsD9 is both thermolabile and
PAAT,
whereasthe DNA polymerase activity ofa re-vertant,under the same conditions, is
thermo-stable andPAA8.
Analysisofintertypic recombinants between HSV-1 and HSV-2 has proved to be a precise methodfor the
physical mapping
of HSVgenetic markers, including a paar mutation (16) andvirus-induced polypeptides (10, 14-16, 19, 24,
29). Inparticular,
intertypic
markerrescue has beenhighlydevelopedas a meansofphysically
mappinggenetic loci (24).In this communication we describe the precise physical mapping by intertypic marker rescue ofpaar
mutations of HSV-1 and HSV-2 and two ts mutations, tsH(HSV-1)and ts6(HSV-2).Mutants
carrying
the tsH and ts6 markers have been shownpreviouslytoinducethermolabileDNA
polymerase
activi-ties (5; I. K. Crombie, Ph.D.thesis,
University
ofGlasgow, Glasgow,Scotland, 1975).
MATERIALS AND METHODS
Cells andviruses.Babyhamsterkidney (BHK-21
265
on November 10, 2019 by guest
http://jvi.asm.org/
266 CHARTRAND ET AL.
C13) cellsweregrown inEaglemediumsupplemented
with 10% tryptosephosphate broth and 10% calf serum (11).Cellmonolayers (2 x 106 to 4 x 106cells) in 50-mm-diameter plastic petri dishes(Flow Laboratories, Inc.) were usedthroughout.
The ts mutants of HSV-1 strain 17 (tsH) and of HSV-2 strainHG52 (ts6) have been described
previ-ously (13, 25, 26). The HSV-1 strain 17 (ts+ paar-1) and HSV-2 strain HG52 (ts+ paar-2) mutants were those isolated and described by Hay and
Subak-Sharpe (6).
Virus DNA preparation and production of HSV DNA fragments. Virion DNA was prepared fromcellsinfected at310Catamultiplicity of infection of102 as describedbyWilkie(27).
Thepreparationof restriction endonucleasesXbaI,
HindIII,EcoRI,BglII, andHpaIand the conditions used for thecleavageofHSV-1 and HSV-2 DNA were asdescribedpreviously (2, 28).
Intertypic marker rescue and analysis of
re-combinants. We havepreviouslydescribed in detail the technique for intertypic marker rescue (24). Briefly, BHK cellsarecoinfectedatthenonpermissive temperaturewith0.4
jig
of intact DNA of a ts mutant of one serotype and0.4jigof unseparated restriction endonucleasefragmentsof DNAfrom ts+ virus of the other serotype.DNA infectionsarecarriedoutbythe Stow and Wilkie (23) modification of the technique of Graham and Van der Eb (4). ts+ virus progeny from such crosses areplaque purifiedthree times at 38.5°C, and the parental origin of restriction endonuclease sites inthe DNA is determined (19, 24). The rationale behind theanalysisof such recombinants is that DNA sequences from the ts+ parent carrying a ts+ gene sequenceshouldreplacethe DNA sequences ofthets parentatthe site of the ts lesion. In the present study, the DNAfrom thets+ parents carried the nonselected marker(paar)for resistance to 100jigof PAA perml. ts+ recombinantsweretestedfor the presence of this markerby determiningtheirrelativeefficiencyofpla-quing(EOP)in the presence andabsence of thedrug at38.5and31°C.
Therecombinants described in this studywereall derived fromindependent crosses and were therefore knowntobeclonallyunrelated. Recombinants R6-17, R6-18, and R6-19, which wereisolated from three
well-separatedplaquesfrom the same dish and could have been related, nevertheless proved to have different genomestructures (seeFig.7).
Physicalmaps of HSV-1 and HSV-2. Figure 1 shows the physical maps ofHSV-1 (strain 17) and HSV-2 (strain HG52) for the restriction endonucleases
XbaI,HindIII, EcoRI,BglII,HpaI, KpnI, andBamHI
in theregion 30 to 50 map units of the genome (24; A. Davison,personal communication). One map unit is equivalentto 106daltons (1.7 kilobase pairs) and the genome orientation isasdescribedpreviously (24, 29). Each map indicates theposition of the restriction sites and corresponding fragments, above for HSV-1 and belowfor HSV-2. At the top of Fig. 1 is a summary of all the restrictionsites combined from the seven maps. The numberscorrespond to the numbers given to each restrictionendonuclease, and the sites are aligned with thecorrespondingonesfrom each map. The order of these sites for eachserotype was deduced by
recleav-30
1-XbaI
2-Hind III
3-Eco RI
4-Bgl II
5-HpaI
6-KpnI
7-Bam HI
35 40 45 50
4 5 5 4 15
3
331
36 6 6 6 6 6
i7,l7177 717 777 7
7 77 7 77
4 5 5 4 2 4 1
e
c d
a
h e
0 C
t b h
III
j d
a h d
m t n p v a'x c
I
q m t 9 d
p u de g v r kw ib d
In I I
nn'I' b rh'j a yI j
FIG. 1. Physical maps of HSV-1 (strain 17) and HSV-2(strain HG52) for seven restriction endonucle-ases(see text).
ageofindividualfragments (3,28; A.Davison,personal communication). Thealignmentof HSV-1 restriction sites with respect to HSV-2 restriction sites is the result of DNA/DNA hybridization experiments (N. M.Wilkieand R.Cortini,unpublished data)andfrom thestudyof the genomestructureof therecombinants
presentedinthis paper. Thisalignmentcannot,
there-fore, be considered to be completely accurate, al-though thealignedmapsshown inFig.1areconsistent with all of the recombinant structures presented
herein.
RESULTS
Phenotypic characterization of inter-typic recombinants. In markerrescue
experi-ments in which PAA is usedas a selection for recombinants,wehavefoundahighbackground
ofPAAr virus,whichmakesmappingvery diffi-cult (N. D. Stow, Ph.D. thesis, University of Glasgow, Glasgow,Scotland, 1978).Forthis
rea-son, thepaarmutationsweremappedas
nonse-lected markers in recombinants produced by intertypiccrossesbetween intact tsPAA8virus DNA andunseparated restriction fragmentsof ts' PAAr virus DNA, using ability to grow at
38.50Castheselectivepressure.
Tables 1and 2list the 40recombinants ana-J.VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.504.259.450.55.367.2]MAPPING OFpaar MUTATIONS OF HSV-1 AND HSV-2 267
lyzed
inthisstudy,
give
the restrictionendonu-cleases used to
produce fragments
of the ts+DNA,
and show therelative EOP38.5°C/31°C
and the relative EOP in the presence and
ab-sence ofPAA for each recombinant and their
parents.
Therelative EOP38.5°C/31°C
valuesranged
from 0.4 to5.0,
indicating
that every recombinantwasts+.
The relative EOP±drug
values for
PAAN
recombinantsranged
from 0.2to
1.7,
whereasPAAN
recombinants gave values from 10-4 to10-3.
The relative EOP ±drug
determinations shown in Tables 1 and 2 were
carried out at
310C,
but similar values wereobtainedat
38.5°C.
Restrictionendonuclease
mapping
ofin-tertypic
recombinants.
The method ofdeter-mining
theparental origin
of DNA sequencesinintertypic
recombinantsby
restriction endonu-cleaseanalysis
has been describedpreviously
(19, 24, 29).
Three factors must be considered wnenmterpreung
t.of DNAfromther(
detected
provided
TABLE 1. Intertyj
ts+ DNA Recombi- fragments
nant produced by: R6-3 HindIII R6-4 HindIII R6-5 HindlIl R6-6 HindIII R6-7 HindlIl R6-8 XbaI R6-9 XbaI R6-10 XbaI R6-11 XbaI
R6-13 HpaI R6-14 HpaI R6-15 HpaI R6-17 EcoRI
R6-18 EcoRI R6-19 EcoRI R6-20 EcoRI R6-21 EcoRI
R6-22 EcoRI R6-23 EcoRI
R6-24 EcoRI
R6-26 BglII R6-27 BglII
R6-28 BglII R6-29 BglII R6-30 BglII R6-31 BgIIl R6-32 BglIl R6-33 BglII
R6-34 BglII R6-35 BglII
HSV-2
(ts6 paa') HSV-1
(ts+ paa-1)
'DNA Cross:HSV-1 paa');selection:ts+ (abi
hEOPin thepresencE
31°C;similarresults at3
TABLE 2. Intertypic recombinants: RHseries'
tsR
DNA Titerat Relative Relative Recombi- fragments 310C EOPEP±nant produced (PFU/ml) 381.5C/ drug'
by: 10
RH-11 HindIII 3.2x10# 5.0 0.4
RH-12 HindIII 2.5 x108 1.8 0.5
RH-13 HindIII 2.7x108 3.3 0.4 RH-14 HindIII 4.2 x1i0 2.4 0.8
RH-21 HpaI 5.2x108 2.5 0.2 RH-22 HpaI 1.2x108 3.9 0.2 RH-23 HpaI 4.9 x107 2.2 0.4 RH-24 HpaI 9.6 x107 2.5 0.2
RH-31 EcoRI 1.6x107 3.8 0.3 RH-32 EcoRI 9.7x106 3.3 0.3 HSV-1 1.6x107 1.3x10-4 <lo-4
(tsH paa')
HSV-2 2.0xi05 5 0.5
(ts+ paa'-2)
'DNA cross: HSV-1 (tsH paa')x HSV-2 fragments (ts' paa'-2); selection:ts+ (ability to growat38.5°C).
bEOP in the presence (100ug/ml) and absence of PAA at 31°C; similar resultsat38.5°C.
ne
resuits.
rirst,
mepresence clease profile of the putative recombinant ge-escuing fragment willonly be nome is altered from that of the parental ge-that the restriction endonu- nome. Second, definition of the length and po-sition of the inserted rescuingsequences dependspicrecombinants: R6seriesa onthe
alignment
of thephysical
maps for HSV-Titerat Relative Relative 1 and HSV-2 DNA (see above). Third, in cases 31°C EOP EOP± in which the size of the recombinant genome is (PFU/ml)38.5oc/
drug' unaltered andonly loss ofrestriction endonucle-1.0X i09 0.5 0. ase sites can be found, weassume replacement 1.9x109 0.9 0.5 ofgenomicsequencesof the mutantby rescuing 1.3 x 109 1.2 1.6 DNA sequences at thosesites.54
xo1081.2
1.7
Figures 2,
3, 4, and 5 show the restriction 9.9 xlo8 1.3 1.3 endonucleasefragmentpatternsof recombinants5.7x10 0.8 1.1 in the RH and R6 series. The RH series was
1.9x109 0.8 0.8
5.9x108 1.1 1.0 isolated after the rescue of HSV-1 tsH DNA 6.6x108 1.0 1.7 withDNA fragments of HSV-2 (ts+
paar-2),
and 1.4x109 0.6 1.1 the R6 series was isolated after the rescue of1.1 x109 1.0 <10-4 2 ts6 DNA with DNA fragments of
HSV-2.6 xio8 0.5 0.7+
7.7 x
108
0.5 0.5 1(ts'
paar_1).
In theinterest
ofbrevity,
only 1.1 x 109 0.9 0.5 those key recombinantswhich define the limits813
x10"117
0.24
x10;l ofmapping
areshown. Theanalysis
ispresented
6.3x108 1.3 0.7x10-' bydiscussingthe restrictionenzyme mapsof the 9.6x108 0.4 0.3 R6 and RH groups enzyme by enzyme, to show
1.2x
10"
0.8 0.2x10-4
that the same limited region of the genome is 1.0x109 1.3 1.3 affected in each case. Changes in restriction7.6x108 0.6 0.9
3.8x108 1.2 1.1
°x
10-l
enzymesitesoutside thisregion
willnotbe dis-6.4x108 0.8 1.1 cussed indetail.To aid theanalysis,opencircles1.2x
i8
0.5 0.5 indicate the loss of fragments compared with the6.6x
108
1.7 0.6 ts parents, and solid squares indicate the ap-1.4x109 0.6 0.7 pearance of new fragments not present in the ts2.8x108
<10-_5
1.1Xl0-4
parents. Restriction sites will bedesignated by
the two letters that identify the fragment on 8.6x
108
1.1 0.8 either side(Fig.
1).
From the data shown in
Fig.
2, it can be deduced thatR6-9, R6-14,and R6-30 havelost fragments(tspaa'-1)
xHSV-2(ts6 the HSV-2 Hindlll h-e site. This produces ailit(y100
g/l)andabsenceofPAA at fusion fragment which migrates slightlyslower38.5°C. thanHSV-2HindlIl a.R6-26showsno change
VOL. 31,1979
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.504.263.456.79.241.2]268 CHARTRAND ET AL.
HSV1 R6-26 HSV-2
R6 4 R614 R6-30
RH-1i HSV-2
HSV1 H-12
_ _
s
._ij~~~~~-
._ . ,.ik:_
J,~~~~~~~~~~~~~~~~~~~~~~
#j: _ _Cdt...
.}~~~~O
w tJ_.==~~~~1=
.11i
0
---C
Hind III Hirid III
RH 14 HSV-2
HS;V PH 22
*_ C~~ad
cdefg 3 ". I-1
.4.
-U1
b-.a
cde....f
hi<_m
LCU R
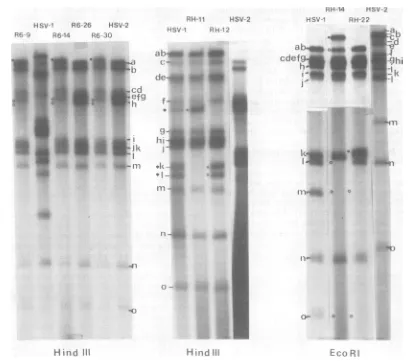
FIG. 2. Autoradiographs of32P-labeledDNAofintertypic recombinants and their parents digested with HindIII andEcoRI. TheEcoRIgelisacompositeofdifferentexposuresofthesamegel.Theparentalpatterns
arefrom infections with the actual parents usedtogenerate therecombinants, i.e., HSV-1 (ts+paa'-1)and HSV-2 (ts6paa')forthe R6 series andHSV-2(ts+ paar-2) and HSV-1 (tsHpaa')forthe RHseries. DNA fragmentsareletteredaspreviously (24). Designations:U,newfragments found in recombinants whichare notpresentin thetsparent;0,fragments foundintsparentswhicharemissinginrecombinants;*,fragments
duetosegregationofmixtures present in theparental stockofHSV-1 tsH.
compared with HSV-2ts6. The stock of HSV-1 tsH used consists ofamixedpopulationinwhich
approximatelyhalf the molecules have lost the HindIIIk-isitetogenerate a fusion band which migrates fasterthanHindIII f (bandsdesignated with an asterisk). Cloned recombinants have segregatedinto oneorthe other oftheseforms, and RH-11 and RH-12 show no other change in their HindIIIpatterncompared with the HSV-1tsHparent. The EcoRIanalysesof RH-14 and RH-22 show the lossof the HSV-1EcoRI m-o site, givingrise tolarger fragments.
Figure3showstheanalysiswithBglII. R6-9, R6-14, and R6-26 share the loss of the HSV-2 BglIIofragment. R6-14hasalsolostthe HSV-2BglII j fragment, but this is not the case for
R6-9 and R6-26. It therefore follows that R6-9 and R6-26 have lost the HSV-2 BglII o-csite. R6-30 has a profile similar to R6-9 (data not shown). RH-li and RH-12 show the reverse situation,having acquiredthe HSV-2BglIIo-c siteandconsequentlyasmaller fragment replac-ing the normal HSV-1 BglII d fragment. Al-though notclearon the gel shown, RH-14 has the sameBglIIrestrictionprofileasRH-11 and RH-12. RH-22 haslost theHSV-1 BglII i
frag-mentand hasacquiredtwo newfragmentswith themobilityof HSV-2BglII jand o.
The KpnIanalysisinFig.4showsthatR6-9, R6-14, and R6-30have allacquireda new
frag-mentwith themobilityofHSV-1KpnIv.These recombinants have also acquired a fragment
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.504.46.454.67.428.2]MAPPING OF paa' MUTATIONS OF HSV-1 AND HSV-2 269
HSV-1 R6-26
R6-9 R6-14 HSV-2
RN-" HSV-2
HSv_l RH-12
RHI4 HSV-2
HSV-1 RH-22
_ l--~b a.
o~cde C.
Z f d.
e.
f
- h g
hl
ij
_
k..
1
1
n 0
o -o p
4-#4 4
aI
a
Bgl 11
Bgl 11
Bgl
11FIG. 3. Autoradiographs of 32P-labeledDNAof intertypic recombinants and their parentsdigested with BglII.Fordetails,seethelegendtoFig.2.
which has themobility of HSV-1 KpnIpinthe case of R6-14 and a slightly lower mobility in
thecasesof R6-9and R6-30. This is because all
three recombinants have acquired the HSV-1 KpnIp-vsite, butonly R6-14 has alsoacquired
the HSV-1 KpnI n-p site. R6-26, RH-11, and
RH-12 show no change compared with theirts
parents. The important feature in RH-14 and
RH-22 is the acquisition of a new fragment
which migrates slower than HSV-1 KpnI abcd duetothe lossoftheHSV-1KpnIx-csite.
Theanalysis withBamHI is shown in Fig.5;
in this case the structure of the recombinant
which carries the nonselected paar_l marker (R6-9) and those which are sensitive to PAA (R6-14, R6-26, and R6-30) canbecompared. All
oftheserecombinants have lost HSV-2 BamHI h', but only R6-9 has also lost the adjoining
f
fragment. The three PAA' recombinants have therefore retained the HSV-2 BamHIh'-f sitebut lost the b-h' site. It follows that thepaa-i
mutation must lie to the right of the HSV-2 .*;A...ia_
qXp
i.
u 0
VOL. 31,1979
_P
_q
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.504.63.451.67.525.2]270 CHARTRAND ET
ALo
abc
cl ef
RH-11 HSV-2 RH-14 HSV-2
HSV-I RH-12 HSV-1 RH-22
Sm
_
-.~~~~~~
rs
::i'XA 4 :i...I...x
It
FIG. 4. Autoradiographs of 32P-labeledDNAof intertypicrecombinants and their parentsdigested with
KpnI. Shown are composites ofdifferent exposuresofthe samegels. The asterisk indicates incompletely digestedDNA. Fordetails,seethelegendtoFig.2.
BamHI b-h' site andcanbe separated fromthe HSV-1ts+counterpartof thets6mutation.The sameregionisinvolvedinthe RHrecombinants. RH-11andRH-12bothhave lostHSV-1BamHI r (difficultto see since rcomigrates with q in this HSV-1 stock), and a new band which mi-grates slower than HSV-1 BamHI b' has ap-peared due to the acquisition of the HSV-2 BamHI b-h' site.Furthermore,RH-12 contains anew fragmentwhich comigrates with HSV-2 BamHIh', and theinsertion of HSV-2 DNA in RH-12therefore includesthe HSV-2BamHI
h'-f'
site. RH-14 and RH-22 show changes in the same region, but in addition have acquiredlongersegments of the HSV-2 genome.
By inspection of therestriction mapsshown in Fig. 1, it can be concluded that all of the changesinrestriction sites foundinthegenomes of R6-9, R6-14, R6-26, R6-30,
RH-li,
RH-12, RH-14, and RH-22 are totally consistent with thegenomestructuresshown inFig.6, 7, and8. The genome structures of the other recombi-nants werededucedinthesameway.Analysisofintertypicrecombinants. The genome structuresindicatingtheparental origin
of the restriction endonuclease sites for seven
enzymes for 35recombinants areshownin Fig.
6, 7,and8.Figure6showsrecombinants isolated from cells coinfected with intact HSV-2 (ts6
paas) and unseparated HindII, XbaI, or HpaI J. VIROL.
imikvciL
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.504.60.453.77.477.2]MAPPING OF paa' MUTATIONS OF HSV-1 AND HSV-2 271
HSV-1 R6-14 R6-26
RH-14 R6-9 HSV-2
up
_
to-4w
w
.
4mR6-30
RH-11 HSV-2
HSV-1 R H-12 RH-22
_.-,~-w_i
l_
ow.-d ~ ~ s-li
S:i
p-
qr-0
0 0
s -
--
--
...-*X nseF *; w
j
IV- __
_
L w
W-
__
__
_.
r_
_ _:"
....
._ *
rsW
b- Z i jVPvE
0
0.XO
-.d,s. ^Bl#i
^£ M_ .iffeE;ifa
,3i: m :. ...
r -- -> ss-. s;
t
e.
S--e-
4 -11
- sie*X_ _ X .s
'F . w ''
s.,'5s'. ,. .:
.... j.
' , _
_3;.:.
BamHI
BamHI
FIG. 5. Autoradiographs of32P-labeledDNA of intertypic recombinants and their parents digestedwith BamHI.Shown arecomposites ofdifferent gels. DNA fragmentsidentifiedbyA.Davison (personal commu-nication). Fordetails,seethelegendtoFig.2.
fragments of HSV-1(ts'paar-1)DNA. Allofthe is compared with those of the other recombi-recombinants containHSV-1DNAsequences in nantsshown inFig.6, itcanbe concluded that theregion betweenmapunits38and41.Eleven thepaar_1 lesionmustlietotherightof thets+ of the12recombinantsare
phenotypically PAAr,
(HSV-1) counterpart of ts6 (HSV-2).indicatingstronglinkage betweenpaar_1and the Inanattempttoobtainfurtherrecombinants ts+(HSV-1)counterpartofts6(HSV-2). Recom- which
split
thesesites,
a second markerrescuebinant R6-14 is PAA8 and, assuming that it is
experiment
was carriedoutusing
either EcoRI notsimplyaPAA revertantofthepaar-1 (HSV- orBglII fragmentsofHSV-1 (ts+ paa _1)DNA. 1)mutation,inthis recombinantpaa'-i
and the EcoRI andBglII
have sites within theregion
of ts+ (HSV-1) counterpart of ts6 (HSV-2) have HSV-1 DNAbetweenmapunits38and41(en-beensplit.When thegenome structure of R6-14 zymes 3 and 4,
respectively,
inFig. 1).
TheVOL. 31,1979
a
sA.
on November 10, 2019 by guest
http://jvi.asm.org/
272 CHARTRAND ET AL.
30 35 40 45 50
4 5 5
6 6
7I177
6 6
4 15
?I I 11 I
3
747'777 77
313
4 5 5 4 214 1
67 6
5 5
6
6 3 5
6
6 1. 3 5
67 3 5
76 76 6
m m
~~~~~~-7
67 7
6 6
6 7
67 6
7
7 6
4 7
7 6
76 6
7
6
izzzIzI~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~zzzzIzIz~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
counterpartof thets6 lesionmustmap nearthat site.Four other isolates from thiscross (R6-17,
R6-21, R6-22 and R6-23[Table 1]) were
pheno-typicallyts+andPAASbut containedonly HSV-2 restriction endonuclease sites with the seven enzymesused. Inthis casetheywere probably
ts+ revertants of HSV-2 ts6,or they contained
undetected insertionsofHSV-1 DNA.
Recombinants R6-26, R6-27, R6-28, R6-29,
R6-4 R
R6-30,
R6-31, R6-32, R6-33, R6-34,
and R6-35wererescued with HSV-1 BglII fragments (Fig.
7). Onlyoneisolate(R6-27,Table1) had neither
R6-6 R detectable HSV-1 restriction sites present nor
evidence of HSV-2 sitesmissing;alloftheothers
R6-7 R contained overlapping HSV-1 DNAsequences.
Seven of the nine recombinantscontained
HSV-R6-5 R 30 35 40 45 50
R6-15 R
R6-4 R
R6-11 R
R6-13 R
R6-3 R
R6-9 R
R6-10R
R6-14 S
FIG. 6. Summaryofthegenomestructureof inde-pendent recombinants isolated from the cross be-tweenintactHSV-2(ts6paa)DNA andunseparated restrictionfragments ofHSV-1
(ts'paa'-1)
DNA. Foreachgenomestructure, the HSV-2 DNA sequences
are shown in whiteand the HSV-1 DNAsequence3 areshown inblack,with theregion of uncertainty of theposition ofthecrossovershownas ahatchedarea.
The numbersindicate thedelimitingrestriction sites (above fortypeIand belowfor type 2).The codefor thesenumbers is thesame as inFig. 1. Rindicates PAA',andS indicatesPAX.
genome structures of recombinants obtained from this experiment areshown in Fig. 7.
Re-combinantsR6-18, R6-19, R6-20, and R6-24were
rescued with HSV-1 EcoRI fragments and are
all PAAr. However, in each case the inserted
HSV-1 DNAsequencesspantheHSV-1 EcoRI
site atapproximately mapunit40. The results indicate a strong selection for recombination eventsinvolving HSV-1 DNAfragmentswhich
retainedthisEcoRIsite ("partials" orannealed
ends) and furthersuggest that thets+ (HSV-1)
45 5
3
116 6
1171 777.
4
6 6
7 7
15
3 3 3 6Il 776. 7717
711717
i45 5 4 24 1
3
67
31
66
66
6 7
6 6
6 7
6
6
4
6
6 6
4
6
6 6
4 6
7 6
27
6
4 3x
7
4 3
7/,-7
R6-18 R
R6-20 R
R6-24 R
R6-19 R
R6-33 R
R6-32 R
R6-29 R
R6-31 R
R6-35 R
R6-28 R
R6-34 R
R6-30 S
R6-26 S
FIG. 7. SeelegendtoFig.6.
J. VIROL.
I
3 6
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.504.63.246.55.435.2] [image:8.504.264.455.150.661.2]MAPPING OFpaa' MUTATIONS OF HSV-1 AND HSV-2 273 30
15
L5
61,l
35 40 45 50
5
6
71
14 15
|7l7lll7
, -
W77
1L1 I7
7
74 5 5 4 2 4 1
3
5 3
6 7
5 4
5 4
6
5 4 6
73
74 7
6 6
2 7 7
271.7
3
[image:9.504.57.247.50.431.2]mziizMM
FIG. 8. Summnaryof thegenomestructureof inde-pendent recombinants isolated from the cross
be-tweenintactHSV-1(tsHpaa") DNA andunseparated
restrictionfragments ofHSV-2(ts'paar-2)DNA.For details,seethelegendtoFig.6.
1DNAsequenceswhichspannedtheBgl site
near map unit 40,which suggests that the
ts'
(HSV-1) counterpartof the ts6 lesion alsomaps
nearthis site.Tworecombinants(R6-26and
R6-34) and HSV-1 DNAsequences which did not
contain thissite, showing thatthe ts+ (HSV-1) counterpart of the ts6 lesion mustmap to the rightofit.Tworecombinants(R6-26andR6-30) werephenotypically PAA' and,unless reversion
of thepadr- mutation is in eachcasethe expla-nation, theserecombinants haveseparated the
paar-l site and thets+ (HSV-1) counterpart of
the ts6 lesion. The genome structures of these
PAA recombinants compared with the PAAr recombinants showthatpad-1 must lie to the
right,atasimilarpositionobtainedby
compar-ing R6-14 (PAAX) with the otherPAAr
recom-binants.
The reciprocal expenment was then carried 6 6
71
6 7
out using the mutant HSV-1 tsH (HSV-1 tsH
doesnotcomplement HSV-2 ts6; L.
L.
Messer, Ph.D. thesis, University ofGlasgow, Glasgow, Scotland, 1978). Recombinants were isolated from cells coinfected with intact DNA from HSV-1 (tsHpaa5)
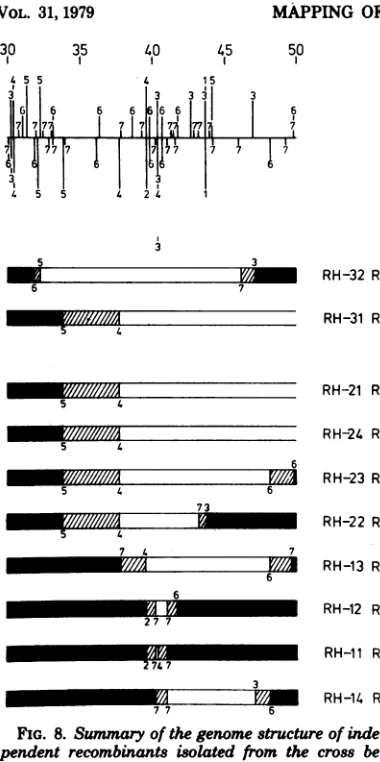
and restriction endonucleasefragments
ofHSV-2 (ts+ paar-2) DNA. The ge-nome structures of these recombinants are shown in Fig. 8. All of the recombinants are PAAr indicating strong linkagebetweenpaar-2and the
ts'
(HSV-2) counterpart of the tsH lesion. Two recombinants (RH-31 and RH-32) wererescuedwithHSV-2EcoRIfiagments butnevertheless contained the HSV-2 EcoRI site near mapunit40,suggesting thatthe ts+ (HSV-2)counterpartoftsH maps near this site. When theuncertainty in the locationof the crossover
points istakeninto account,allofthe recombi-nants of the RH series had HSV-2 DNA se-quencesm commonwithin the HSV-2 BamHI h'fragment.
Summary
ofanalysis.
In theanalysis
shownin Fig. 6, 7,and 8, we have mapped the HSV-1 DNA sequences (or HSV-2 DNA sequences) which correct the ts lesion in HSV-2 ts6 (or
HSV-1 tsH). We assume that the correction
mechanismusually involvesthereplacementof
defective sequences in the tsparent by the ts+ counterpartfrom the other serotype. Other
ex-planationsarepossible, but if the assumption is correct,the map location of the ts6 mutation is
defined by the HSV-1 DNA sequences which the R6series ofrecombinantshave in common.
Inspection of the data shown in Fig. 6 and 7
indicatesthat thisregion is defined onthe left
by the HSV-2 HindIII h-e site or the HSV-1
Bgll
i-dsite(R6-26andR6-34) andontherightby the HSV-2 BamHI h'-j' site (R6-14, R6-26, and R6-30; see Fig. 6, 7, and 1). The region of the genome containing the tsH lesion can be
derived in the same way by inspection of the datafortheRH series(Fig. 8)and is definedby
the HSV-2 BamHI h'fragment,which isa
frag-mentof 1.3kilobasepairs. These data are sum-marized in Fig. 9; note that the limits shown take intoaccounttheuncertaintyinthe location of thecrossoverpoints.
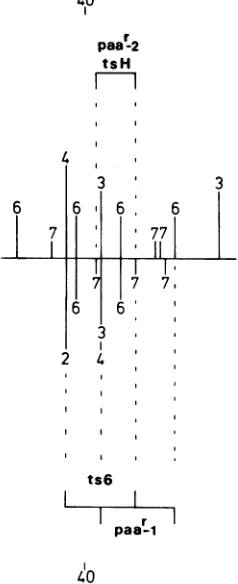
Figure9 also
sumarizes
themaximum
mapcoordinates for the HSV-1 (paa
-1)
and theHSV-2
(paar-2) paar
mutations. Allof the RHseries ofrecombinants acquired PAArand
pre-sumably
contained thepaar-2 mutation. Thus,the map coordinates which define thepaar-2 mutation are the same as those for the tsH lesion. Three R6 recombinantswerePAA',and therefore
thepaa-1
mutationwassplit
fromthe ts' (HSV-1) counterpart ofts6. Comparison of the genome structureofR6-14,R6-26,
andR6-VOL. 31,1979
3
on November 10, 2019 by guest
http://jvi.asm.org/
274 CHARTRAND ET AL.
30with the remainingrecombinants in the R6 series showsthatpaar_1 mustmapattheright
of ts6 between the coordinates shown inFig.9. These are defined on the left by the HSV-2 BgIII o-c site and onthe right bythe HSV-1
KpnIx-csite(see Fig.6, 7, and1).
DISCUSSION
This communication describes the detailed physical mapping of mutations (tsH and ts6)
present in ts mutants of HSV-1 and HSV-2 which inducethermolabile DNA
polymerase
ac-tivity (5; Crombie, Ph.D. thesis) and those (paar_1 andpaar-2) present intwomutants re-sistant to PAA. Bothphenotypesareconsistent with lesions in genes which affect viral DNApolymerase activity insome way.The method of mapping involved genetic analysis of
inter-typic recombinants combined with restriction endonuclease mapping of their DNA. The
ra-tionale behindsuchanapproachhasbeen stated
previously (19, 24; thispaper), but it is
worth-40
6
7 6
paat2 tsH
I I
3, 3
6,.
3
6 771
111
II I
17
7 7,6 6 ' 3,
24,3
Il
ts6
r
paa-I
[image:10.504.96.215.315.607.2]40
FIG. 9. Summary ofthemappingdatafor tsH,ts6,
paar_1, andpaad-2. Theregion ofthegenomeshown
isanenlargement ofthe 39- to 43-map unitregion shown inFig. 1.
while to consider in more detailwhat is actually being mapped in such experiments. The inter-pretation of mapping results for ts lesions is based on the assumption that rescuing DNA sequences contain the ts+ counterpart of the ts mutation and thattheyreplace sequences at the site ofthe mutation. Thisinterpretationwould be invalid if "second site" rearrangements re-sulting in inter- or intragenic suppression were commonly responsible for the observed pheno-types of the recombinants. The recombinants isolated in this study show a wide range in the length of rescuing DNA sequences, which argues against crossovers whichresult inintragenic sup-pression of the mutantphenotype. Another gen-eral drawbackto the mapping rationalewould be in cases in whichheterozygosity ispossible. Inthis case ats+ phenotype couldbeacquired
withoutloss of the gene coding for the ts phe-notype. In the present work, thereciprocal map-ping of HSV-1 and HSV-2 mutations whichlie
in the same complementation group (tsH and ts6
[Messer,
Ph.D.thesis])
puts both mutations in aregion of DNA smaller than that thought necessarytocode for theexpectedprotein(DNApolymerase [see below]). Since intratypic marker rescue experiments using isogenic ma-terial also put both mutations into the same region (P. Chartrand, N. M. Wilkie, and M. C.
Timbury, manuscript in preparation; Stow,
Ph.D. thesis), this reduces the likelihood that
heterozygosityisaproblem forthislocus.
Inter-pretation of the mapping of paar (the nonse-lectedmarker) depends onlyontheassumption that inserted DNA sequences contain the paar mutations.
Given that the assumptions are correct, the
mapping studies with tsH and ts6 delimit the locationfor both thetslesions.Theclones which contain inserted HSV-1 DNA sequencesallow mapping of thepaar_1 (HSV-1) mutation, and those with HSV-2 DNA insertions allow the mapping of the paar-2 (HSV-2)mutation.From
Fig.9itcanbeseenthat all four mutations map in one region of HSV DNA, which spans ap-proximately2 x106daltons (3.3 kilobase pairs). When intratypic marker rescue (isogenic sys-tem) was used, the mapping of tsH and ts6 mutations gave very similar results (Chartrand
et al., manuscript in preparation; Stow, Ph.D. thesis).
This is the first report of the simultaneous and independent mapping of an identifiable function of HSV-1 andHSV-2 (PAA'),and the resultsstrongly suggest closelysimilar map lo-cations. Theuncertaintyinthefinealignmentof the HSV-1 and HSV-2physical maps may ob-viously affect theinterpretation,butit should be
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
MAPPING OF
paa'
MUTATIONS OF HSV-1 AND HSV-2 275 noted that themaplocations for the mutationstsH, ts6, and paar-2 are defined by restriction
endonuclease sites in the HSV-2 physicalmap.
Recent studiesutilizingHSV-1/HSV-2 inter-typicrecombinants havepermitted the mapping of several HSV-induced polypeptides (14, 16).
The order ofpolypeptides in the region under study (14) was found tobe Vmw 117 (glycopro-tein), Vmw 136'(130) (DNA bindingprotein), and Vmw28. In the Marsden etal. (14) study, five
intertypic recombinants isolatedafterrescueof
HSV-1 tsHwereanalyzed. The onlypolypeptide
whichneversegregated fromthe rescuing DNA
sequences wasthe Vmw 136'(130) DNA binding
protein. This probably excludes the genes for
theotherpolypeptides from containing the site forthetsHmutation, butit doesnotprovethat
the gene for the V,w 136'(130) protein does
contain the site for the tsH lesion.Purified viral DNApolymerase has beenreported tohave a
molecularweightof150,000(18), but it hasnot yetbeenassigneda maplocus.
Sincemutantscarrying the tsH,ts6,paa-r1,or
paar-2 mutation show phenotypic properties compatible with lesions in virus-induced DNA polymerase activity (5, 6; Crombie, Ph.D. the-sis), thequestion arises whether thefour lesions
are inthe same gene. tsH andts6do not
com-plement each other (Messer, Ph.D. thesis) and, provided unforeseencomplications donotarise inintertypic complementation tests, these
mu-tationsarelikelytoreside in thesame gene.The paa-r andpaar-2 mutations lie very near the
tsHandts6 lesions. Thecodingcapacity needed for the 150,000-molecular-weight polypeptide whichPowelland Purifoy (18) found ina
puri-fied preparation of HSV DNA polymerase is approximately4.6kilobasepairs.The maximum mapping limits (3.3 kilobase pairs; Fig. 9) are
thereforeconsistentwithallfourmutationslying
inthestructuralgeneofthe viral DNA polym-erase,but donotprovethathypothesis. A paar mutation hasrecently been mapped (9), using intratypicmarkerrescue,andreportedtolieto
theright of the HSV-1HpaI b-h site (map unit 45;seeFig. 1). This isatleast3.8kilobasepairs
tothe right of thepaar lesionsreported in this
study. This raises the possibility thattwo inde-pendentgenescancarrymutations which result
inaPAAF phenotype. If the PAArphenotype is
the result of modifications in the viral DNA
polymerase, this mayimplytwoloci whichare
involved in DNA polymerase activity. In this respectit is interesting to notethattwots
mu-tations(tsC4 andtsD9) which lieintwodifferent HSV-1 complementation groups (22) induce a
viralDNApolymeraseactivity which, when
ex-tensively purified,isthermolabile in vitro (20; K.
L.Powell and D.J. M. Purifoy, personal
com-munication).ThetsD9mutation, which also
re-sultsin a PAA'phenotype(seeabove),has been found to map between 0.30 and0.45 (fractional
length)(D. S. Parris and P.A.Schaffer, personal communication). Although this is consistent with our mapping data, more precise physical mappingand furthercomplementationtestswill berequiredtoresolve theprecise relationshipof the differentpaar mutations and the tsH, ts6,
tsD9,andtsC4lesions.
ACKNOWLEDGMENTS
WethankM.MurphyandC.Adair for cheerful and expert assistance, A. Davison for thepreparationof restriction en-donucleases, and J. Hayforproviding seed stocksof the PAA' mutants.Wealso thankK.L.Powell, D. J.M.Purifoy,D.S. Parris,and P. A. Schaffer for thepersonalcommunication of unpublished data.WearegratefultoJ. H.Subak-Sharpefor his interestthroughoutthisstudy andforvaluablecriticism of themanuscript.
P.C. is therecipientofafellowship from the Canadian MedicalResearchCouncil.
LITERATURE CITED
1. Aron,G. M., D.J.M. Purifoy,andP. A.Schaffer. 1975.DNAsynthesis andDNApolymeraseactivityof herpessimplexvirus type1temperature-sensitive mu-tants.J.Virol.16:498-507.
2. Clements,J.B.,R.Cortini,and N. M.Wilkie. 1976.
Analysis ofherpesvirusDNA substructurebymeansof restriction endonucleases.J.Gen.Virol.30:243-256. 3. Cortini, R., andN.M.Wilkie.1978.Physicalmaps for
HSV type 2 DNA with fiverestriction endonucleases. J.Gen. Virol. 39:259-280.
4.Graham, F. L,andA. L.Van der Eb. 1973. Anew techniquefor the assay ofinfectivityof human adeno-virus 5 DNA.Virology 52:456-467.
5. Hay,J.,H.Moss,A.T.Jamieson,andM.C.Timbury.
1976.Herpesvirusproteins:DNApolymeraseand py-rimidinedeoxynucleoside kinaseactivitiesin tempera-ture-sensitivemutantsofherpessimplexvirus type2.
J.Gen. Virol.31:65-73.
6. Hay, J., andJ. H. Subak-Sharpe. 1976. Mutants of herpessimplex virustype1and2thatareresistantto phosphonoacetic acidinducealteredDNApolymerase activities ininfectedcells. J.Gen.Virol. 31:145-148.
7. Honess,R.W.,andD.H.Watson.1977.Herpessimplex virusresistance andsensitivitytophosphonoaceticacid. J. Virol. 21:584-600.
8. Joffre,J.T.,P.A.Schaffer, and D.S.Pan-is. 1977.
Geneticsof resistancetophosphonoaceticacid in strain KOSofherpessimplexvirus type1. J.Virol. 23:833-836.
9. Knipe,D.M.,W. T.Ruyechan,andB. Roizman. 1979. Molecular geneticsof herpessimplexvirus. III. Fine mapping ofageneticlocusdetermining resistanceto phosphonoacetatebytwomethodsofmarkertransfer. J. Virol.29:698-704.
10. Knipe,D.M.,W. T.Ruyechan,B.Roizman,andL.W. Halliburton. 1978.Moleculargeneticsofherpes sim-plexvirus.Demonstration ofregionsofobligatoryand non-obligatory identity within diploidregions of the
genome. Proc. Natl.Acad. Sci. U.S.A.75:3896-3900. 11.Macpherson, I., and M. Stoker.1962.Polyoma
trans-formation ofhamstercell clones-aninvestigation of geneticfactorsaffectingcellcompetence.Virology16:
147-151.
VOL. 31, 1979
on November 10, 2019 by guest
http://jvi.asm.org/
276 CHARTRAND ET AL.
12.Mao,J.C. H., E. E.Robishaw,and L. R.Overby.1975.
Inhibition of DNA polymerase activity from herpes simplex virus-infectedW-38cellsby phosphonoacetic acid. J. Virol. 15:1281-1283.
13. Marsden, H. S.,I. K. Crombie, and J. H.
Subak-Sharpe. 1976. Control of protein synthesisin herpes-virus-infectedcells:analysis of thepolypeptide induced
bywild type and sixteentemperature-sensitivemutants
of HSVstrain 17. J. Gen.Virol.31:347-372.
14.Marsden, H. S., N. D. Stow, V. G. Preston,M. C.
Timbury, andN. M.Wilkie. 1978.Physical mapping ofherpes simplex virus-induced polypeptides.J.Virol.
28:624-655.
15.Morse, L S., T.G.Buchman,B.Roizman,and P. A.
Schaffer. 1977.Anatomyofherpes simplexvimsDNA. IX.Apparent exclusionofsomeparentalDNA
arrange-mentsin thegenerationofintertypic (HSV-1xHSV-2)
recombinants.J.Virol.24:231-248.
16. Morse, L S., L. Pereira, B. Roizman, and P. A. Schaf-fer. 1978. Anatomy ofherpes simplex virus (HSV) DNA. X.Mapping of viralgenesby analysisof polypep-tides and functions specified byHSV-1xHSV-2
recom-binants. J.Virol.26:389-410.
17. Overby, L. R.,E. E.Robishaw,J. B.Schleicher,A.
Ruter,N.L.Shipkowitz, and J. C. H. Mao. 1974.
Phosphonoacetic acid:inhibitor ofherpes simplexvirus.
Antimicrob.AgentsChemother. 6:360-365.
18. Powell, K. L, and D. J. M.Purifoy.1977.Nonstructural proteins ofherpes simplex virus.I.Purification of the induced DNApolymerase. J. Virol. 24:618-626.
19. Preston, V. G.,A.J.Davison,H.S.Marsden, M.C.
Timbury,J. H.Subak-Sharpe,and N. M.Wilkie. 1978.Recombinantsbetweenherpes simplexvirustypes
1and 2:analysisofgenomestructuresandexpression of immediateearly polypeptides.J. Virol.28:499-517.
20. Purifoy,D. J.M.,R. B.Lewis,and K.L.Powell.1977.
Identification of the herpes simplex virus DNA
poly-merasegene.Nature(London)269:621-623.
21. Purifoy, D. J. M., and K. L Powell. 1977. Herpes simplex virus DNA polymerase as the site of phospho-noacetatesensitivity: temperature-sensitive mutants. J. Virol. 29:470-477.
22. Schaffer, P. A., G. M. Aron, N.Biswal, and M. Ben-yesh-Melnick. 1973. Temperature-sensitive mutants of herpessimplex virus type 1:isolation, complementation and partialcharacterization. Virology 52:57-71. 23. Stow, N. D., and N. M. Wilkie. 1976. An improved
technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J.Gen. Virol. 33: 447-458.
24. Stow, N. D., and N. M. Wilkie. 1978.Physical mapping oftemperature-sensitive mutations ofherpessimplex virus type 1byintertypicmarkerrescue.Virology90:
1-11.
25. Timbury, M. C. 1971.Temperature-sensitivemutantsof herpessimplex virus type 2. J. Gen. Virol. 13:373-376. 26. Timbury, M. C., and L Calder. 1976. Temperature-sensitive mutants of herpes simplex virus type 2: a provisional linkage map based on recombination anal-ysis. J. Gen.Virol. 30:179-186.
27. Willie, N. M. 1973. The synthesis andsubstructure of herpesvirus DNA: thedistribution of alkali-labile single strandinterruptionsinHSV-1DNA. J. Gen.Virol.21: 453467.
28. Wilkie, N. M. 1976. Physical maps for herpessimplex virus type 1DNA forrestriction endonucleases Hind III,Hpa-1and X. bad. J. Virol. 20:222-233.
29. Wilkie, N. M., N. D.Stow,H. S.Marsden,V. Preston, R. Cortini, M. C. Timbury, and J. H. Subak-Sharpe.1977.Physicalmapping of herpes simplex virus coded functions andpolypeptidesbymarker rescue and analysisofHSV-1/HSV-2intertypic recombinants, p. 11-31. InG. de The, W. Henle, and F. Rapp (ed.), Proceedings of the Symposium onHerpesvirusesand Oncogenesis.III, part 1. International Agency for Re-search Against Cancer, Lyon, France.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/